More Information
Submitted: August 25, 2023 | Approved: September 01, 2023 | Published: September 04, 2023
How to cite this article: Wang GG, Zhang Y, Wang XY, Zhang GL. Microbial Conversion and Utilization of CO2. Ann Civil Environ Eng. 2023; 7: 045-060.
DOI: 10.29328/journal.acee.1001055
Copyright License: © 2023 Wang GG, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: CO2 fixation; Bioconversion; Microorganisms; Value-added products; Metabolic engineering
Microbial Conversion and Utilization of CO2
Ge-Ge Wang1#, Yuan Zhang2,3#, Xiao-Yan Wang2,3* and Gen-Lin Zhang1
1School of Chemistry and Chemical Engineering, Shihezi University, China
2Nutrition & Health Research Institute, China National Cereals, Oils and Foostuffs Corporation (COFCO), China
3Beijing Key Laboratory of Nutrition, Health and Food Safety, China
#These authors contributed equally to this work and should be considered co-first authors
*Address for Correspondence: Xiao-Yan Wang, Nutrition & Health Research Institute, China National Cereals, Oils and Foodstuffs Corporation (COFCO), Beijing, china, Email: [email protected]
Rising greenhouse gas emissions have contributed to unprecedented levels of climate change, while microbial conversion and utilization of CO2 is a practical way to reduce emissions and promote green manufacturing. This article mainly summarizes several natural CO2 pathways that have been discovered, including the Calvin cycle, the reduced tricarboxylic acid (rTCA) cycle, the Wood–Ljungdahl (WL) pathway, the 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle, the dicarboxylate/4-hydroxybutyrate (DC/HB) cycle, the 3-hydroxypropionate (3HP) cycle, the reductive glycine (rGly) pathway, and artificially designed carbon fixation pathways includes the CETCH cycle, the MOG pathway, the acetyl-CoA bicycle, and the POAP cycle. We also discussed applications of different carbon fixation enzymes, notably ribulose-1, 5-diphosphate carboxylase/oxygenase, pyruvate carboxylase, carbonic anhydrase, as well as formate dehydrogenase. This paper further addressed the development of photosynthetic autotrophs, chemergic autotrophs and model bacteria Escherichia coli or yeast produced main products for CO2 fixation through metabolic engineering, such as alcohols, organic acids, fatty acids and lipids, bioplastics, terpenoids, hydrocarbons, and biomass. Future studies on CO2 microbial conversion should focus on improving the efficiency of carbon fixation enzymes, metabolic modules of the carbon sequestration pathway, and intracellular energy utilization. Coupled microbial and electrochemical methods for CO2 fixation, in addition to biological fixation, show considerable promise.
Global temperatures have reportedly risen by 1 ℃ as a result of human activity and natural events, including ocean sinks, earthquakes, and volcanic activities, since the beginning of the Industrial Revolution. The rapidly increasing CO2 concentration in the atmosphere has contributed to a number of environmental problems, including global warming, a decline in biodiversity, ocean acidification, and even modifications to industrial production methods and human lives. If growth continues at the current pace, the global temperature will continue to rise, worsening extreme weather, ecological disasters, and other negative effects [1]. In order to alleviate climate stress, 178 countries signed the "Paris Agreement" in 2016. They promise to limit the average global temperature increase this century to 1.5 ℃. By 2030, carbon emissions will need to be reduced globally by 40% in order to achieve that objective.
Carbon capture, utilization and storage (CCUS) technologies use net-carbon or low-carbon energy sources to achieve the purpose of reducing carbon dioxide on a worldwide scale [2,3]. CCUS, which develops an environment that is both sustainable and ecologically friendly, represents a different approach to achieving reduction goals [4]. CCUS technology consists of several steps: CO2 capture, transportation, utilization, and storage. First CO2 is absorbed and separated through physical [5] and chemical absorbers [6], metal-organic frameworks (MOFs) [7,8], ionic liquids [9], bioenergy with carbon capture and storage (BECCS) [10], membranes [11], etc. The CO2 is then transported to a place of storage or a location for carbon use. Lastly, the captured CO2 is subsequently employed or stored as resources using engineering techniques. The main method that CO2 is used as a resource includes direct use (such as the production of dry ice, refrigerants, CO2 microbubble technology [12,13], chemical conversion (such as the production of high-value fuels, chemicals, building materials, and minerals) [14,15], electrochemical catalytic conversion of CO2 [16,17], thermal catalysis [18], photocatalysis [19], enzymatic conversion [20], coupling electrochemical CO2 conversion with CO2 capture [21], microbial utilization [22] and coupling photo/electrocatalytic with microbial CO2 utilization [23]. Among these, the technology of microbial CO2 utilization and conversion is endowed with particular advantages including mild reaction conditions, minimal pollution, low cost, and low energy consumption, which is supposed to be a promising way of reducing carbon emissions [24,25].
In this paper, we will mainly discuss the development of microbial CO2 utilization and conversion technology. This review concentrates on both natural and artificial CO2 fixation pathways, as well as the use of key CO2 fixation enzymes like ribulose-1,5-diphosphate carboxylase/oxygenase (RuBisCo), formate dehydrogenase (FDH), pyruvate carboxylase (PC), and carbonic anhydrase (CA), and the advancement of photosynthetic autotrophs, chemoautotrophs, and model bacteria like E.coli and yeast in fixing CO2 through metabolic engineering, genetic engineering, and synthetic biology. The existing limitations and challenges associated with the biological conversion and utilization of CO2 are then described.
Carbon dioxide fixing pathways
According to features like ATP usage, carbon fixation reactions, enzymes involved, and carbon species being fixed, the natural carbon fixation pathways have been divided into the Calvin-Benson-Bassham (CBB) cycle, the reductive tricarboxylic acid (rTCA) cycle, the Wood-Ljungdahl (WL) pathway, the 3-hydroxyproppionate (3HP) cycle, the 3-hydroxyproppionate/4-hydroxybutyrate (HP/HB) cycle, the dicarboxylate/4- hydroxybutyrate (DC/HB) cycle and reductive glycine (rGly) pathway. In recent years, scientists have synthesized several in vitro CO2 fixation routes based on the kinetics of enzyme reactions, including the CETCH cycle [26], the MOG pathway [27], the ACB cycle [28] and the POAP cycle [29] (Table 1) . In the following, we will present these pathways and discuss their reaction and progress.
| Table 1: Characteristics of pathways for CO2 fixation. | ||||||
| Pathway | Key enzymes | Carbon source | Product | ATP | NAD(P)H | Ref |
| CBB cycle | Ribulose-1,5-diphosphate carboxylase/oxygenase Phosphoribulokinase |
3 mol CO2 | GA-3P | 9 mol | 6 mol | [30] |
| rTCA cycle | ATP-citrate lyase 2-ketoglutarate synthase |
2 mol CO2 | Acetyl-CoA | 2 mol | 4 mol | [36] |
| W-L pathway | Formate dehydrogenase CO dehydrogenase Formylmethanofuran dehydrogenase |
2 mol CO2 | Acetyl-CoA | 1 mol | 4 mol | [39] |
| DC/HB cycle | 4-Hydroxy butyryl-CoA dehydratase | 1 mol CO2 + 1 mol HCO3- | Acetyl-CoA | 3 mol | 4 mol | [45] |
| HP/HB cycle | 4-Hydroxy butyryl-CoA dehydratase | 2 mol HCO3- | Acetyl-CoA | 4 mol | 4 mol | [42] |
| 3HP cycle | Malonyl-CoA reductase Malyl-CoA lyase |
3 mol HCO3- | Pyruvate | 5 mol | 5 mol | [47] |
| rGly cycle | Reductive glycine cleavage complex | 1 mol CO2 | Pyruvate | 2 mol | 3 mol | [48] |
| CETCH cycle | Crotonyl-CoA carboxylase/reductase | 1 mol CO2 | Glyoxylate | 1 mol | 7 mol | [26] |
| MOG pathway | Phosphoenolpyruvate carboxylase | CO2 | Glyoxylate | 8-12 mol | 6 mol | [27] |
| ACB cycle | Ferredoxin oxidoreductase | 2 mol CO2 | Acetyl-CoA | 5 mol | 5 mol | [28] |
| POAP cycle | Pyruvate carboxylase Acetate-CoA ligase Oxaloacetate acetylhydrolase Pyruvate:ferredoxin oxidoreductase |
2 mol CO2 | Oxalate | 2 mol | 1 mol | [29] |
Natural CO2 fixation pathway
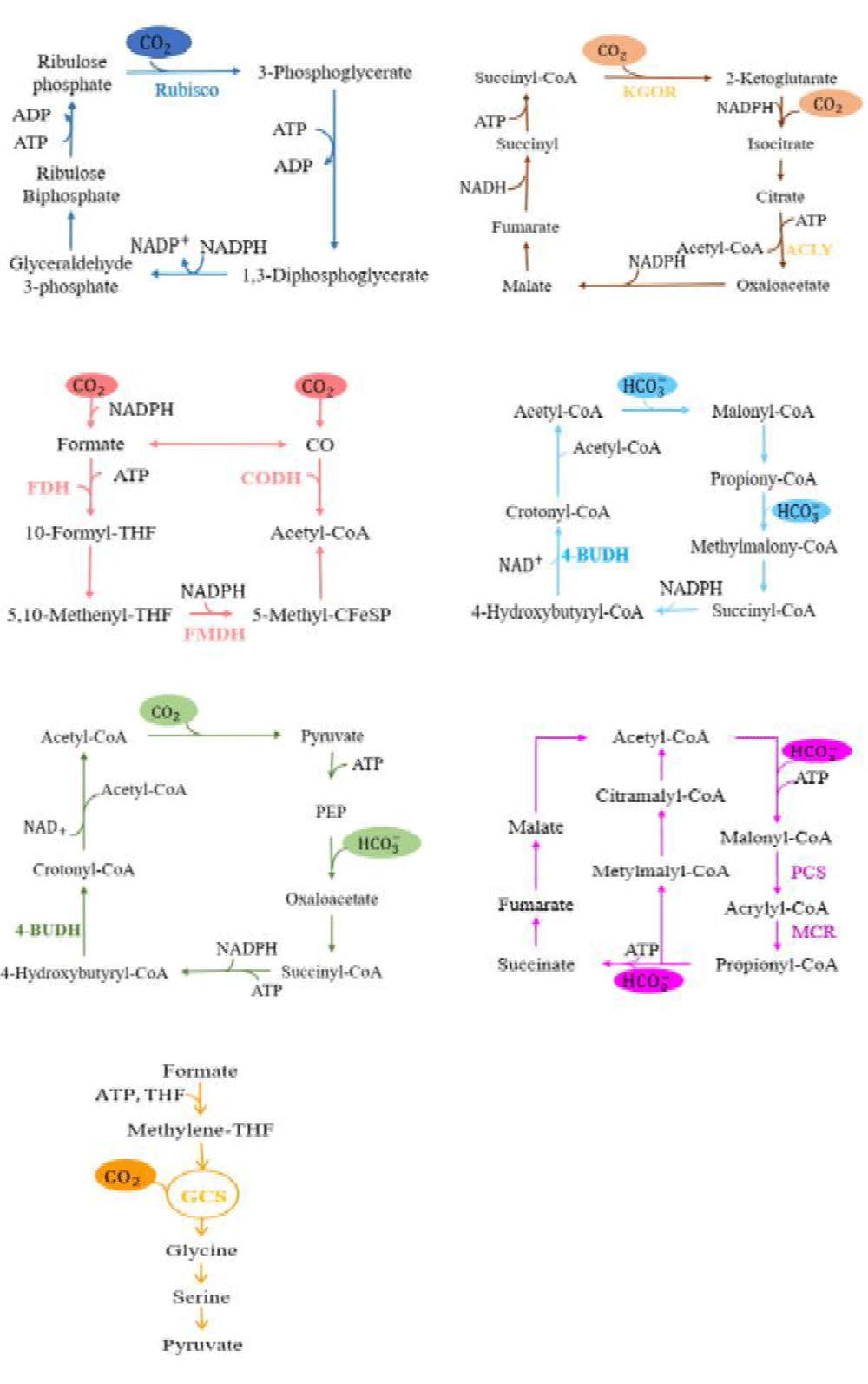
Calvin-Benson-Bassham cycle: The majority of photosynthetic plants, algae, and proteobacteria use the Calvin-Benson-Bassham cycle (CBB cycle) as their primary route for fixing CO2 in nature. There are 13 enzymatic processes, of which ribulose-1, 5-diphosphate carboxylase/oxygenase, and phosphoribulokinase are the major enzymes. This cycle can be divided into three stages: (1) carboxylation, also known as CO2 fixation, a molecule of CO2 is integrated into a five-carbon compound called 1,5-diphosphate ribulose (RuBP), resulting in an unstable C6 compound that breaks down into two molecules of a three-carbon compound known as 3-phosphoglyceric acid; (2) reduction, NADPH reduces 3-phosphoglyceric acid to Glyceraldehyde 3-phosphate while consuming ATP; (3) regeneration, glyceraldehyde 3-phosphate through a series of reactions to produce ribulose diphosphate [30].
Researchers have successfully introduced the CO2 fixation process of the Calvin cycle into model organisms like Escherichia coli and yeast using genetic engineering techniques. This development overcomes the limits of photoautotrophic carbon sequestration and enables heterotrophic organisms to progress fully into autotrophy utilizing CO2 as the only carbon source [31]. By integrating the CO2 fixation pathway with energy consumption pathways in E. coli, Gleizer and colleagues [32] created a fully autotrophic E. coli strain that can thrive only on CO2. Gassler and coworkers controlled the endogenous gene expression in Pichia pastori to deactivate a portion native methanol metabolism pathway. They then added a heterotrophic CBB cycle for carbon fixation, which transformed Pichia pastoris from a heterotrophic to an autotrophic organism capable of utilizing CO2 for growth [33].
Methanol is a C1 substrate that methylobacterium uses as a source of biomass and energy. Methylobacterium extorquens AM1 was engineered by Borzyskowski and group to carry out both energy metabolism and biomass synthesis, with energy coming from methanol metabolism and biomass synthesis deriving from CO2 metabolism via the Calvin cycle [34].
Through the CBB cycle, photosynthetic autotrophs naturally fix about 300 billion tons of carbon dioxide each year. The rate of carbon fixation is a key factor limiting plant development in environments under conditions of abundant water and light. To increase the metabolic flow of the CBB cycle, CO2-fixing enzyme efficiency must be enhanced [35].
Reductive tricarboxylic acid cycle
The reductive tricarboxylic acid (rTCA) cycle has since been found in Thiobacillus viridis, bacteria, and archaea. In this cycle (Figure 1) , the enzyme ATP-citrate lyase breaks down citrate into acetyl-CoA and oxaloacetic acid. Malate dehydrogenase then catalyzes the enzymatic reduction of oxaloacetic acid to malic acid, which generates fumaric acid through fumaric hydration. Fumaric acid is further transformed into succinic acid through the enzyme reductase. Finally, succinic acid generates citrate by redox [36].
Malubhoy and coworkers engineered yeast strains to produce succinic acid (SA) from pyruvate by rTCA cycle, the greatest yield of SA was 0.23 Cmol/Cmol glycerol [37]. Kang and colleagues overexpressed pyruvate carboxylase, malate carboxylase, and malate transporter simultaneously adding xylose metabolic pathways and knocking out the natural yeast pathways that create ethanol and glycerol. Through the rTCA cycle, this allowed engineered yeast to produce malic acid [38].
WL pathway
The WL pathway, also known as the reductive acetyl-CoA pathway, was discovered by Wood, Ljung-dahl, and others, and exists in acetonogenes, acetogenes, and some fungi. During one of the two branches, one CO2 molecule is reduced to a single methyl group, while in the other branch, one molecule of CO2 is reduced to CO, which then cooperates with the methyl group and CoA to generate acetyl-CoA. Acetyl-CoA can be further converted into biomass or acetyl phosphate. Subsequently, the acetyl combines with ADP to produce ATP and acetate [39]. CO dehydrogenase (CODH), formate dehydrogenase, and formylmethanofuran dehydrogenase are the main enzymes in this pathway (Figure 1).
Figure 1: Natural CO2 fixation pathways. the CBB cycle (in blue) [30]; the reductive TCA cycle (in brown) [36]; the Wood–Ljungdahl pathway (in pink) [39]; the HP/HB cycle (in cyan) [42]; the DC/HB cycle (in green) [44]; the 3-HP cycle (in purple) [47]; the reductive glycine pathway (in orange) [48]. RuBisCo: Ribulose-1, 5-diphosphate Carboxylase/oxygenase; ACLY: ATP-Citrate Lyase; KGOR: 2-Ketoglutarate ferrioxoreductase; FDH: Formate Dehydrogenase; CODH: CO Dehydrogenase; FMDH: Formylmethanofuran Dehydrogenase; 4-BUDH: 4-Hydroxy butyryl-CoA dehydratase; PCS: Propionyl CoA synthase; MCR: Malonyl-CoA Reductase; GCS: the Glycine Cleavage System.
Papoutsakis and colleagues expressed 11 enzymes and auxiliary core protein genes from Clostridium in Clostridium acetobutylicum demonstrating that the two branches can work independently of one another. They discovered that acetyl-CoA synthase (ACS) catalyzes the condensation of CO with methyl for the synthesis of acetyl-CoA, whereas CODH catalyzes the reduction of CO2 to CO. They also learned that CODH/ACS forms complexes that connect two branches of the WL route [40]. By introducing ACS and ACDH into E. coli, Hu, and colleagues from Jiangnan University created a novel CO2 fixation metabolic route (HWLS). They combined the fixation and utilization of carbon dioxide in two modules, producing butyrate and malic acid with yields of 1.48 mol/mol glucose and 0.79 mol/mol glucose, respectively [41].
HP/HB cycle
The autotrophic thermococcus utilizes acetyl-CoA/propionyl-CoA carboxylase as the primary carboxylase for CO2 fixation, according to research by Berg and colleagues [42]. Through the reduction of 3-hydroxypropionic acid, one molecule of acetyl-CoA and two molecules of bicarbonate are transformed into succinyl-CoA in this system. Succinyl-CoA is then reduced to 4-hydroxybutyric acid, which is subsequently converted into two molecules of acetyl-CoA by 4-hydroxybutyryl CoA dehydratase (Figure 1). Recycling bicarbonate within cells represents a more feasible evolutionary pathway than CO2. In Pyrococcus furiosus, Keller and team effectively produced 3-hydroxypropionic acid by overexpressing five HP/HB cycle genes derived from Metallosphaera sedula [43].
DC/HB cycle
The dicarboxylic acid/4-hydroxybutyric acid (DC/HB) cycle exists in anaerobes and facultative aerobes, such as Thermoproteales and Pyrolobus fumarii [44]. One molecule of CO2 and one molecule of bicarbonate are fixed by pyruvate synthase and phosphoenolpyruvate (PEP) carboxylase in this pathway, respectively, to produce succinyl-CoA [45]. The DC/HB cycle has not been successfully expressed heterologously because it requires unique iron, sulphur, and thioester proteins, and other things.
3HP cycle
Through the acetyl-CoA/propargyl-CoA carboxylase, the 3-hydroxypropionate (3HP) cycle effectively integrates two moles of bicarbonate [46]. The four steps in this metabolic process are as follows: (1)propionyl CoA synthesis from acetyl-CoA; (2) propionyl CoA conversion to succinate; (3) glyoxylate production; and (4) glyoxalate and propionyl CoA assimilation reactions to generate pyruvate followed with acetyl CoA regeneration. Through acetyl-CoA/propargyl-CoA carboxylase, the 3-HP cycle assimilates 2 moles of bicarbonate [47]. The enzyme of this cycle has noteworthy characteristics such as effective CO2 fixation, oxygen insensitivity, abundant intermediates, and the capacity to regenerate glycolic acid and glyoxylic acid. In E. coli K12, Mattozzi and colleagues expressed genes in an operon linked to the 3-HP pathway from various species. They successfully set up a heterologous 3-HP metabolic pathway and evaluated its functionality via a growth curve. The findings show that the four subpathways are all capable of functional heterologous expression [47].
Reductive glycine pathway
Anaerobic bacteria use the reductive glycine (rGly) route to assimilate formate. Tetrahydrofolate, a cofactor in this pathway, activates formate into formyl-tetrahydrofolate while expending one molecule of ATP in the process. Then, methylene tetrahydrofolate is produced by further reducing formyl-tetrahydrofolate. After the condensation and reduction of methylene tetrahydrofolate by CO2, NH3, and NADH, glycine is produced. The reversible glycine cleavage system (rGCS), which consists of the aminomethyltransferase T protein, glycine decarboxylase P protein, dihydrolipoamide dehydrogenase L protein, and the aminomethyl carrier H protein, is the main component of rGlyP [48,49].
Bang and colleagues expressed tetrahydrofolate (THF) cycle and formate dehydrogenase in E. coli to create a formic acid utilization route, which allowed the engineered strain to grow without the need for additional glucose supplementation [50,51]. Cruz and coworkers overexpressed the enzyme MIS1 together with other enzymes involved in the glycine cleavage/synthesis system in yeast, establishing a glycine synthesis route that allows yeast to generate glycine from formate and CO2 [52]. The reductive glycine pathway was modularized by Kim and colleagues into four parts (C1, C2, C3, and energy), and these modules as well as methanol dehydrogenase were introduced into E. coli. As a result, C1 substrates such as methanol, formic acid, and CO2 were effectively used by modified bacteria for growth [53].
The rGly pathway, along with the WL pathway and TCA cycle, is one of the most efficient ATP pathways among the verified CO2 fixation pathways. As a result, under anaerobic industrial circumstances, the reductive glycine pathway has become an incredibly attractive production method. However, insufficient ATP biosynthesis in anaerobic conditions prevents the production of a variety of compounds via these pathways [48]. In order to produce ATP efficiently and use a variety of single-carbon substrates for sustainable biosynthesis, future research should focus on investigating and improving the glycine synthase system.
Artificial CO2 fixation pathways
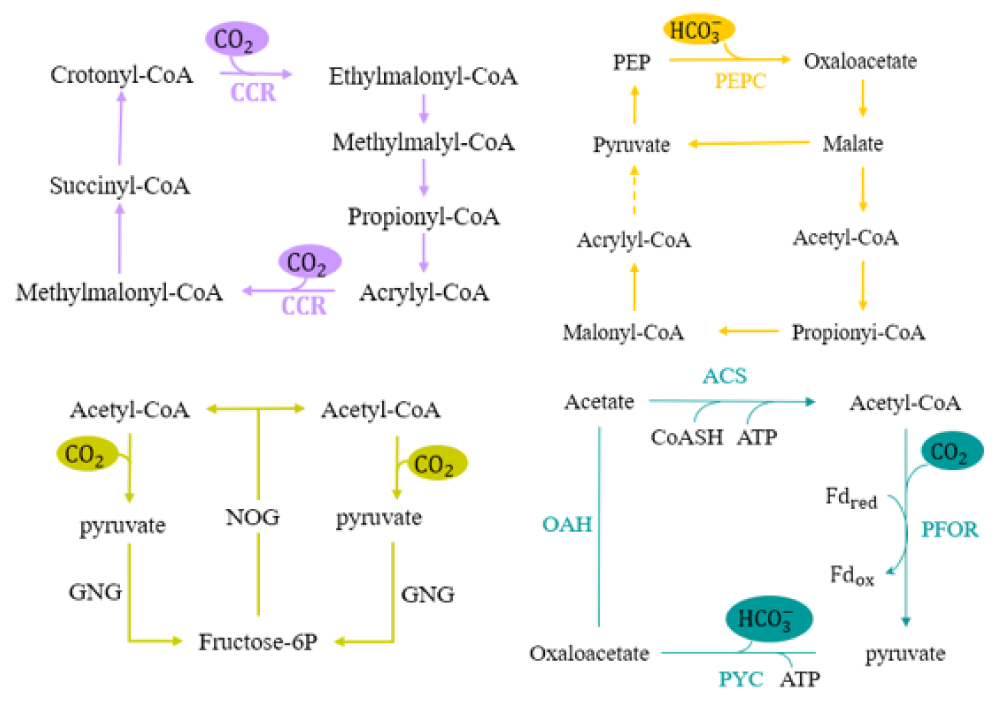
In addition to the natural pathways for CO2 fixation mentioned above, researchers have also created a number of artificial pathways for CO2 fixation depending on features like topology, ATP efficiency, and thermodynamics (Figure 2) .
Figure 2: CCTN Architectural Design Co., Ltd. Oct Jining Grand Canal Culture & Art Center. Architectural Practice. 2023, (02):92-100 [23].
CETCH cycle
The CETCH cycle, which consists of 17 enzymes obtained from nine distinct species including animals, plants, and microbes, indicates the synthetic CO2 fixation route in vitro. The CETCH cycle compared to the naturally occurring CO2 fixation pathway, demonstrates higher kinetic and thermodynamic favorability after multiple optimization through enzyme engineering and metabolic engineering. This cycle overcomes the carboxylation bottleneck in natural carbon fixation pathways by employing highly carboxylated enoyl-CoA carboxylases/reductases (ECRs) as a major enzyme for converting CO2 into organic molecules at a rate of 5 nanomoles per minute per milligram of protein. ECRs exhibit catalytic efficiency 2-4 times better than RuBisCo, the primary carbon fixation enzyme of the CBB cycle. It exists in secondary metabolism, utilizing molecular oxygen as a substrate and not participating in autotrophic carbon fixation pathways. Notably, this new carbon fixation route uses less energy and calls for fewer reaction steps [26].
MOG pathway
The MOG pathway was synthesized by Bar-Even and coworkers by combining the metabolic modules of many organisms. They compared the specific activity and affinities of phosphoenolpyruvate carboxylase, pyruvate carboxylase, acetyl-CoA and propionyl-CoA carboxylase and other carboxylases towards CO2 or HCO3- and metabolic pathways, revealing that these metabolic pathways generate glyoxylate via the same metabolic pathway. Consequently, these pathways were termed the MOG (malonyl-coenzyme A-oxaloacetate glyoxylate) pathways. Notably, the MOG pathways exhibit significant quantitative advantages, such as the overall kinetic rate, cycle rates, and carboxylation efficiency [27].
Acetyl-CoA bicycle
Acetonogenic bacteria metabolize C1 substrates into C2 metabolites, such as acetyl-CoA. Wu, et al. sequentially connected three functional modules—carbon fixation, gluconeogenesis, and non-oxidizing glycolysis, establishing the reductive acetyl-CoA bicycle (ACB) process, a novel CO2 fixation method, by following this metabolic pathway [28]. In the ACB pathway, two molecules of the C1 substrate (CO2 or formic acid) and two molecules of acetyl-CoA are catalyzed by the enzyme pyruvate:ferredoxin oxidoreductase (Pfor), resulting in two molecules of pyruvate. Then, through gluconeogenesis, these pyruvate molecules are converted into one molecule of hexose. Through glycolysis, the hexose is further broken down into three molecules of acetyl-CoA. One of the acetyl-CoA molecules generates a C2 product, the other two molecules reenter the cycle. After one full cycle, two molecules of a C1 chemical generate one molecule of acetyl-CoA. In natural syngas fermentation strain Clostridium ljungdahlii DSM 13528 coexpressing phosphoketolase and the ACB pathway, the engineered bacteria growth rate and carbon fixation efficiency increased under three different culture conditions: gas only, sugar only, and gas-sugar mixture [54].
POAP cycle
The POAP cycle consists of a four-step reaction: (1) pyruvate:ferredoxin oxidoreductase (Pfor) carboxylates acetyl-CoA to pyruvate, which contrary to the natural metabolic pathway, is the most difficult step in the cycle; (2) pyruvate carboxylase (Pyc) then converts pyruvate into oxaloacetic acid through carboxylation; (3) oxaloacetic acid is hydrolyzed by the enzyme oxaloacetate acetylhydrolase (Oah), which releases acetic acid and oxalic acid; (4) acetic acid is converted into acetyl-CoA by the enzyme acetyl-CoA ligase (Acs) [29].
The POAP pathway completes one cycle, the conversion of two molecules of CO2 into one molecule of oxalic acid at the expense of two molecules of ATP, and one molecule of NAD(P)H. This cycle involves the fewest steps among artificial carbon fixation cycles and enables fixing CO2 under anaerobic and higher temperature conditions.
Despite having an advantage over thermodynamics, dynamics, and energy utilization effectiveness, the artificial CO2 fixation pathway still has challenges, such as the complexity of a multi-enzyme system and a reliance on exogenous energy sources, which fall below the level of the standards of industrial biological manufacturing.
The key enzymes for carbon dioxide fixation: Ribulose-1, 5-diphosphate carboxylase/oxygenase
In the CBB cycle, 1, 5-diphosphate carboxylase/oxygenase (RuBisCo) catalyzes the carboxylation of ribulose 1, 5-diphosphate (RuBP) with CO2 to form 3-phosphoglyceric acid (3-PG). RuBisCo also exhibits high sensitivity towards oxygen and can oxidize ribulose 1, 5-diphosphate to generate 2-phosphoglycolic acid (2-PG) [55]. Based on its structural variations, RuBisCo can be classified into four forms, forms I, II, III, and IV. Form I RuBisCo consists of eight large subunits and eight small subunits. The presence of small subunits enhances CO2 concentration within the environment and large subunit facilitates carboxylation. This structure is termed as L8S8 which is commonly found in eukaryotic and prokaryotic photosynthetic organisms. Form II RuBisCo comprises eight large subunits known as L8 structures primarily observed in spirorubidiums, dinoflagellates, and purple non-sulfur photosynthetic bacteria. Form III RuBisCo enzymes are composed of 2-10 large subunits predominantly existing in archaea or a few bacteria. While form IV, also called the Rubisco-like protein (RLP), does not catalyze either of these reactions. There appear to be six different clades of RLP, mainly present in Bacillus, chlorothiobacillus, and Archaeococcus [56]. Pang, et al. expressed forms I and II of the Rubisco enzyme in E. coli to study how the two forms of the enzyme affect CO2 fixation. They found that the activity of these two enzymes for fixing CO2 was comparable [57].
Fujihashi, et al. designed the mutant SP8-T289D by contrast sequencing RuBisCo from a variety of organisms and introduced it into the mesophilic, photosynthetic bacterium Rhodopseudomonas palustris. As a result, the engineered bacterium displayed an approximately two-fold increase in specific growth rate in comparison to the control bacterium [58]. Aigner, et al. coexpressed the chaperone proteins Cpn60/Cpn20, Raf1/Raf2, RbcX, and bundle-sheath defective-2 (BSD2) with Arabidopsis thaliana RuBisCo in E. coli. They found that BSD2 was crucial for stabilizing the assembly of plant RuBisCo large subunit until the small subunits were available [59].
The ability to fix CO2 has been demonstrated by heterologous expression of the bacterial RuBisCo in yeast, E. coli, and other microbial hosts [60-63]. However, the Rubisco only catalyzes a limited number of molecules per minute, low protein assembly efficiency, energy consumption, carboxylation efficiency, and specific CO2 binding affinity lead to a low carbon fixation efficiency through heterologous expression [64]. Meanwhile, the difficult process of expressing RuBisCo from plants in microbes frequently leads to insufficient carboxylation functionality. In order to increase the effectiveness of heterologous expression in microorganisms, it is vital to explore the carboxylation active site of the RuBisCo and to optimize both carboxylation efficiency and specificity.
Pyruvate carboxylase
Pyruvate carboxylase (PC) catalyzes the carboxylation of pyruvate and HCO3- to form oxaloacetic acid and phosphoenolpyruvate carboxylase (PEPC) catalyzes the carboxylation of phosphoenolpyruvate (PEP) with HCO3- to form oxaloacetic acid and inorganic phosphate. Oxaloacetic acid, a significant TCA cycle intermediate, is used in the biosynthesis of several amino acids, including lysine, threonine, and aspartate. Therefore, the fixation of CO2 by pyruvate carboxylase has great significance for amino acid production [65].
Zelle, et al. expressed pyruvate carboxylase PYC2, malate dehydrogenase (MDH), and malic acid transporters SpMAE1, and the malic acid production in Saccharomyces cerevisiae reached 59 g/L [66]. To further increase fumaric acid production, Xu, et al. knocked down gene fum1 encoding fumarase and overexpressed the fumaric acid transporter and pyruvate carboxylase [67]. Xiberras, et al. constructed the yeast succinic acid biosynthesis pathway and replaced the NAD-dependent dihydroxyacetone pathway with the native glycerol metabolism pathway. The yield of succinic acid produced by modified bacteria under batch culture conditions in glycerol was 10.7 g/L [68].
Carbonic anhydrase
The carbonic anhydrase (CA) is the primary enzyme responsible for the hydration of atmospheric CO2, which has a catalytic conversion frequency of 106 s-1. In addition to CO2 capture, CA also can be used for multi-enzyme-catalyzed conversions, chemical-enzyme-catalyzed conversions, and biological conversions [69] . Interaction with RuBisCo, phosphoribulokinase (PRK), and other enzymes involved in natural CO2 fixation pathways is necessary to enable the CA function in biosynthesis. Introducing CA into E. coli, the flux of the CO2-fixing bypass pathway increased from 13% to 17% [70]. Gleizer et al., coexpressed CA, RuBisCo, PRK, and FDH in E. coli to transform heterotrophic organisms into complete autotrophic organisms through laboratory evolution techniques [32]. Effendi et al., expressed human carbonic anhydrase in E. coli MG1655 using a dual promoter σ70 and heat shock protein (HSP70A) instead of inducers to enhance its activity under high-temperature conditions. Cadaverine was successfully produced by the modified bacteria with a yield of 36.7 g/L using CO2 as a substrate [71].
Formate dehydrogenase
The formate dehydrogenase (FDH), commonly known as CO2 reductase enzyme, catalyzes the transformation of CO2 into formic acid [72,73]. Du, et al. overexpressed yeast formate dehydrogenase in S. cerevisiae, which led to modified yeast that consumed 30% more glucose and produced 13% more ethanol [74]. Wang, et al. combined CO2 fixation and formate consumption in yeast. The modified bacteria formate utilization rate was continuously improved to 0.48 g/L/h, and the FFA titer reached 10.1 g/L under glucose-feeding conditions [75]. Under anaerobic conditions, E. coli produces formate hydrolyzase (FHL), which oxidizes formic acid into CO2 and H2. By raising the pressure of CO2 and H2 gases inside the reactor, Roger, et al. discovered that FHL efficiently converted CO2 and H2 to formate and formate extracellular concentrations accumulated to above 500 mM [76]. Based on FDH's extraordinary efficiency at lowering CO2, it has the potential to directly air capture and use carbon. Formic acid, a byproduct of FHL metabolism, can either be used as an energy source or transformed into other useful substances by chemical or biological processes [77,78].
CO2-fixing microorganisms: CO2-fixing Autotrophs
Autotrophs use CO2 as their main or only source of carbon by photosynthesis or chemosynthesis. It can be achieved to reduce CO2 emissions by modifying autotrophic microorganisms to produce bioproducts while also fixing carbon dioxide.
Photoautotroph
Photosynthetic autotrophic microalgae are able to fix CO2 through their metabolic pathways by using CO2 fixation enzymes like RuBisCo and CA. Microalgae are an excellent resource of biobased feedstock for the generation of biofuels because they have better photosynthesis and a greater CO2 fixation efficiency than terrestrial plants [79]. Microalgae also have advantages in cultivation, growth rate, and oil content. Microalgae are therefore essential for the production of biomass and the fixation of carbon [80,81]. Wei, et al. overexpressed the RuBisCo activator enzyme (nRCA) from Nannochloropsis oceanica in Nannochloropsis spp., which increased the biomass output by 46%, the large subunit protein expression level by 45%, the growth rate by 32%, and the productivity of the lipids by 41% [82]. Wang and Shin, et al. modified the light-trapping antenna protein to increase microalgae photosynthetic system efficiency for solar energy utilization and carbon fixation rate [83,84]. Using 15% CO2 (v/v) as a screening stress and a spotting plate method, Jin, et al. determined that Heynigia riparia SX01 had the highest biomass productivity (0.39 g/L/day) and CO2 fixation rate (0.71 g/L/day) [79]. This study offers insightful information on employing microalgae to convert CO2 from flue gas into biomass feedstock.
Cyanobacteria are another photoautotrophic bacterium that can be easily designed and has low food requirements, similar to microalgae. Researchers have successfully manipulated cyanobacteria to produce alcohols, alkenes, terpenes, and organic acids [85-89]. However, due to their need for light and gas supply, photosynthetic bacteria are only able to produce a limited amount of light-sensitive, volatile, and intracellularly unstable chemicals. Li, et al. proposed an integrated strategy (iPRCC) that combines a carbon sequestration module and a resting cell catalysis module. E. coli was manipulated genetically to transform intermediates into light-sensitive products and intracellularly unstable molecules after modified cyanobacteria were used to drive metabolism toward stable substrates. This study expands the possible use of carbon-negative biosynthesis technology while using CO2 biosynthesis for high-value compounds like vanillin [90].
Along with microalgae and cyanobacteria, purple non-sulfur bacteria like Rhodospiralis and Rhodobacter spheroides, as well as anaerobic photosynthetic purple sulfur bacteria, can utilize carbon dioxide to produce chemicals [91]. Fixen, et al. used a transcription factor NifA mutant to activate nitrozyme expression, regulate intracellular metabolism of electrons and energy, and drive Rhodopseudomonas palustris to convert CO2 into methane [92].
Chemolithoautotrophic
Chemoautotrophs obtain energy by oxidation of environmental electron sources such as ammonia, hydrogen, carbon compounds, and sulfur. The hydrogen-oxidizing bacterium Ralstonia eutropha (also known as Cupriavidus necator) exhibits a wide range of metabolic processes, employing CO2 as the sole carbon source and H2 and O2 gases as substrates [93]. R. eutropha efficiently produces PHB and directs the carbon flux towards biofuels and other high-density carbon chemicals like isobutanol, methyl ketone, isoprene, sucrose, modified PHB, and plant growth accelerator via metabolic engineering techniques to optimize their metabolic pathway [93].
Wang, et al. successfully constructed a glucose metabolic pathway in C. necator H16, by blocking the ED and PHB synthesis routes, enabling hydrogen-oxidizing bacteria to effectively utilise glucose, glycerol, and CO2 for inositol synthesis [94]. Joshua, et al. successfully developed a heterologous (R)-1, 3-butanediol biosynthesis route in C. necator H16 by implementing (R)-3-hydroxybutyraldehyde CoA and pyruvate-dependent pathways, knocking out competing pathways, and increasing butanediol synthesis gene expression levels [95]. Liu, et al. created a novel water-splitting biosynthetic system that enables R. eutropha to synthesize chemicals using hydrogen produced during the water decomposition process under low CO2 concentration. With up to 50% energy efficiency during CO2 reduction, this artificial photosynthetic system demonstrates amazing potential and offers a platform for microorganisms to make use of light energy to fix CO2 [96].
Through the enzyme action of formate dehydrogenase, C. necator is able to convert formates into CO2 and then participate in the Calvin cycle. However, the high ATP need of this process restricts the amount of biomass production. Claassens, et al. established a reductive glycine route in C. necator, achieving similar growth rates compared to the wild type, after short-term evolution, which offers the potential for biologically converting formic acid [97]. Even though R. eutropha has a highly developed genetic system for the bioconversion of carbon dioxide and the capacity to manufacture a variety of chemical products, more research is required to improve its genetic tools and synthetic biology techniques compared to other model microorganisms [98].
Common chemoautotroph Clostridium mainly fixes carbon via the reductive CoA process. Cheng et al., overexpressed aor, adhE2, and fnr in Clostridium carboxidivorans, which increased butanol and ethanol yields by 18% and 22%, respectively [99]. Huang, et al. utilized a phage serine integrase-mediated site-specific genome engineering technique introducing heterologous phage attachment/integration (Att/Int) systems in Clostridium ljungdahlii, the modified strain produced a butyric acid yield of 1.01 g/L [100]. Clostridium ljungdahlii is an anaerobic, non-photosynthetic mixotrophic bacteria, that uses both organic and inorganic compounds, such as sugar and CO2 and H2. Jones et al., genetically modified C. ljungdahlii and the engineered strain acetone output had reached 138% of the theoretical maximum [101].
Additionally, via either the Calvin cycle or the CoA pathway, electroautotrophs can directly or indirectly use electricity as a source of energy for fixing carbon dioxide.
CO2-fixing heterotroph
Natural C1 metabolizing microorganisms can convert CO2 into biofuels and chemicals. However, due to ineffective carbon fixation capacity, which results in significant carbon loss, their bioproduction efficiency is far less than heterotrophs. Industrial model microorganisms have become an attractive host for the construction of third-generation biorefineries in contrast to naturally CO2-fixing microorganisms because of their advanced molecular and synthetic biology tools and well-established fermentation processes [63].
Yeast
Yeast performs exceptionally well in industrial settings, and creating CO2-fixing yeast strains presents an attractive choice for developing carbon-neutral industrial processes. Anaerobic fermentation byproducts like glycerol can be produced due to redox cofactor imbalances. It can increase both carbon use and biological production, and decrease byproduct production by using CO2 produced during fermentation as an electron acceptor for NADH oxidation in microorganisms.
Xia, et al. overexpressed the reductive pentose phosphate pathway in S. cerevisiae SR8 as well as the CO2 fixation enzyme of the CBB cycle to improve xylose fermentation [62]. Prk, RuBisCo, and GroESL were expressed in Saccharomyces IMU032 by Guadalupe, et al. Engineered strain produced 90% less glycerol and 10% more ethanol under conditions of a sugar-limited medium with glucose and galactose fermentation [63]. Li, et al. built the CBB pathway in S. cerevisiae and fermented it in a YP medium supplemented with 70 g/L maltose and 40 g/L xylose, the modified yeast CO2 fixation rate was 336.6–436.3 mg/CO2/L/h [64]. Compared to the metabolism of glucose, the sorbitol metabolism by glycolysis produces one more molecule of NADH. By introducing the sorbitol metabolic pathway into yeast, Van Aalst, et al. increased the efficiency of biological production by providing additional electrons for carbon fixation pathways [102]. Gassler, et al. changed the Pichia pastoris endogenous methanol absorption system into a CO2 fixation pathway through chromosome integration technology. Completely autotrophy strain using CO2 as the only carbon source was successfully obtained after laboratory evolution [33]. Both metabolic engineering and protein engineering should be taken into consideration to optimize Rubisco-PRK pathways and increase protein expression levels in order to achieve functional expression of RuBisCo in yeast; establishing natural mechanisms for intracellular CO2 concentration is also essential for effective carbon sequestration [65].
Yeast fixes carbon dioxide using also the rTCA cycle. Xiberras, et al. integrated expression cassettes for three enzymes that convert oxaloacetate to SA in the cytosol ("SA module") into the yeast genome to produce succinic acid, achieving a maximum yield of 0.22 g/g glycerol [67]. Xu, et al. increased fumaric acid production to 1.6 g/L by metabolic engineering with S. cerevisiae EN.PK2-1C as host [68]. Recently, researchers coupled electrocatalysis and biosynthetic techniques to produce glucose and free fatty acids from CO2 and water, yielding 2.2 g/L of glucose and 448.5 mg/L of free fatty acids [103]. Additionally, expressing pyruvate carboxylase expression in yeast can also synthesize amino acids, ethanol, and other compounds.
Escherichia coli
E. coli is a potential heterotrophic organism to metabolize CO2 due to rapid growth, and abundant genetic tools. Zhuang, et al. successfully expressed RuBisCo and PrkA in E. coli, the engineered strain CO2 emission decreased by 15% compared to the wild strain (JB), while the CO2 fixation rate remained constant at 67 mg CO2/mol arabinose/L/h, similar to microalgae and cyanobacteria [61]. Gong et al., introduced CA into E. coli to enhance intracellular CO2 concentration [70]. Gleizer, et al. coexpressed RuBisCo, Prk, and FDH in modified E. coli to convert heterotrophic bacteria to total autotrophic utilizing CO2 as the only carbon source [32]. When RuBisCo is in the activated state, it catalyzes the conversion of the substrate RuBP and CO2 into two molecules of 3-phosphoglyceric acid (PGA). Pang, et al. coexpressed RuBisCo and Rca in E. coli to study the effect of the activating enzyme on carboxylation activity; which improved host overall metabolism by reducing intracellular RuBP inhibition on the expression of RuBisCo [57]. Hu, et al. overexpressed pyruvate carboxylase in E. coli and the engineered bacteria malic acid production rose by 110% [104]. Bang, et al. constructed recombinant THF and reduced glycine pathways in E. coli, this increased the flux of formic acid and CO2 assimilation towards pyruvate synthesis from 4.5% to 14.9% [50]. Heterologous expression of the carbon fixation pathway allows host assimilation of C1 substrates, but one or multi-step reactions are insufficient for effective CO2 fixation. The efficiency of carbon fixation enzymes may be significantly increased by creating an environment where autotrophs assimilate CO2 in the heterologous host. Prochlorococcus marinus MED4 carboxylase structural protein, stabilizing factor, molecular chaperone, and auxiliary module were coexpressed in E. coli to successfully engineer the synthesis of carboxysomes (CBs), which led to a notable improvement in CO2 assimilation capability [105]. Although the construction of carboxysomes is essential for effective CO2 assimilation, due to the large number of related genes and complicated assembly process, it imposes development expenses. Accelerating CO2 fixation using carboxylases in heterotrophic bacteria requires more research.
Although the conversion of heterotrophic E. coli strains to autotrophic has been accomplished, more study is still required to improve metabolic flux, enhance C1 capture and utilization efficiency, and accelerate the growth of "synthetic autotrophic microorganisms" [106].
Microbial CO2 fixation by metabolic engineering
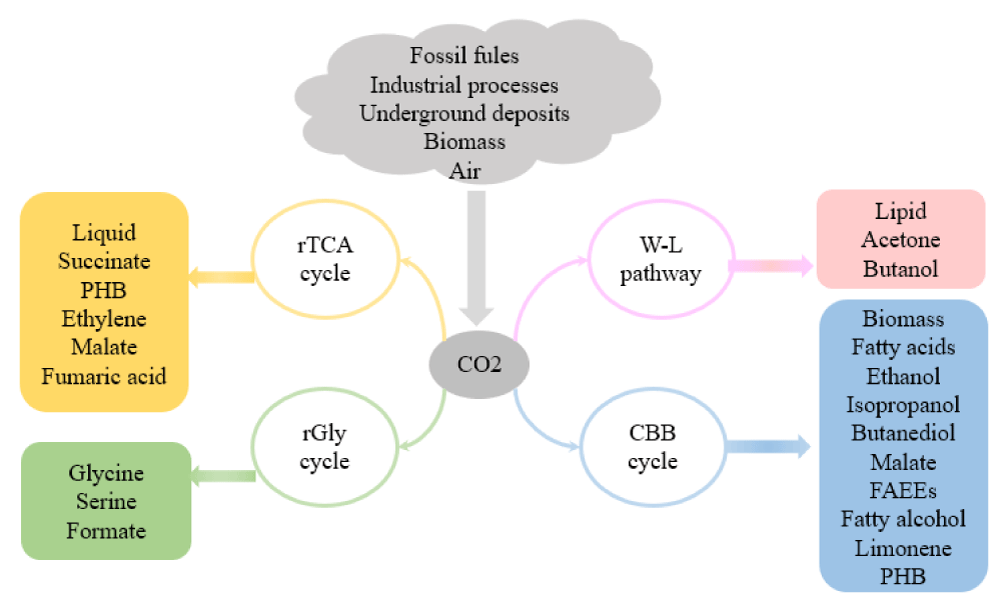
The bioconversion of CO2 exhibits low energy consumption, less pollution, a wide range of products, and high conversion efficiency. Utilizing microbial metabolism to convert CO2 into biobased chemicals is a fundamental strategy in addressing the challenge of increasing global atmospheric CO2 concentration [1]. There are two main ways for microorganisms to metabolize carbon dioxide and produce value-added compounds: one involves building biosynthetic production pathways in naturally carbon fixation organisms, and the other involves converting heterotrophic production strains into "synthetic autotrophic strains" through the use of carbon fixation pathways. This section examines developments in the metabolic engineering of microbial conversion of carbon dioxide into biomass, biofuels, and other important biobased compounds (Figure 3).
Figure 3: Various commodity chemicals and bioproducts derived from CO2 by microorganisms.
Alcohols
Synechococcus and engineered cyanobacteria use carbon dioxide and solar energy to produce alternative fuels or chemicals, which has the potential to greatly reduce reliance on fossil fuels and minimize carbon emissions. Kusakabe et al., successfully engineered a pathway in Synechococcus elongatus PCC7942 for isopropanol synthesis, after optimizing production conditions, the engineered cyanobacteria produced 26.5 mg/L of isopropanol [85]. By deleting the regulatory gene cp12 and overexpressing key enzymes of the oxidative pentose phosphate pathway in S. elongatus PCC7942, Kanno, et al. created engineered bacteria that can produce 12.6 g/L of 2,3-butanediol in both dark and light environments [86]. Shen, et al. modified the S. elongatus PCC7942 by introducing ketoacid decarboxylase, alcohol dehydrogenase, and citramalate pathways to improve the biosynthesis of isoleucine, a precursor to 2-ketobutyrate. The ultimate 2-methyl-1-butanol concentration of the modified bacterium was 200 mg/L, highlighting the first time 2MB was produced through photosynthetic [87]. In Synechocystis sp. PCC 6803, Yao, et al. expressed the fatty acyl-CoA reductase gene maqu_2220 and knocked off competing pathways, which resulted in directed carbon flux towards fatty alcohol synthesis and a final titer reached 2.87 mg/g dry weight [107]. Li, et al. used R. eutropha H16 as the host bacteria and introduced isobutanol and 3-methyl-1-butanol (3MB) synthesis pathways. The modified strain LH74D used formate produced by CO2 electrochemistry as a carbon source with a yield of 0.57 g/liter for 3MB production. This study showed the viability of employing carbon dioxide as a feedstock and electricity as an energy source to drive the biological conversion of carbon dioxide into different compounds [108].
Numerous research have suggested that yeast can make ethanol expressing the heterologous carbon fixation pathway. This strategy reduces the generation of glycerol and increases the output of ethanol by using CO2 as an electron acceptor for NADH reoxidation [109].
Organic acids
Hu, et al. coupled carboxylation reactions that produce ATP in the main metabolic pathway with consume ATP in the natural carbon fixation pathway in E. coli to improve the yield of malic acid [104]. Yu, et al. improved gene expression levels by combining promoters (P4, P17, and P19), CO2 transport genes (sbtA or bicA), and fixed genes (ppc and pck) in E. coli. the modified strain AFP111 compared to the control strain, succinic acid yield increased by 37.5%, reaching 89.4g/L [110]. Kang, et al. overexpressed pyruvate carboxylase, malate carboxylase, and malate transporter in yeast and introduced the xylose fermentation pathway, the resulting strain produced 61.2 g/L of malic acid in fed-batch culture [38]. Wang, et al. successfully enhanced 3-HP biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803 by enhancing the expression levels of essential enzymes, improving precursor supplies of malonyl-CoA and NADPH, and suppressing competing routes. The 3-HP yield of the modified strain reached 837.18 mg/L [111]. D-lactate is essential to produce polylactic acid. The methylglyoxal synthase gene from E. coli was inserted into the cyanobacterium S. elongatus PCC7942 to directly manufacture lactic acid from carbon dioxide through methylglyoxal utilizing dihydroxyacetone phosphate (DHAP). The maximum lactate titer obtained was 13.7 mM (1.23 g/l) [112]. Besides organic acids, microorganisms can also use CO2 to synthesize inorganic acids such as acetate and butyric acid [113].
Fatty acids and lipids
Fuel made from fatty acids is an important biofuel. Microalgae are examples of photosynthetic microorganisms that can use fatty acid synthase to turn CO2 into malonyl-CoA, synthesize fatty acids, and extend the carbon chain. These compounds, such as triglycerides and polyphosphate triglycerides, can then be hydrolyzed for use in the chemical, food, and energy industries [114,115]. Wang, et al. designed a C1 substrate assimilation platform in S. cerevisiae to synthesize free fatty acids (FFAs) from CO2 and formic acid. The formic acid utilization rate increased 21.8 times in the modified strain KW301, while the fatty acid output increased 33.7 times, reaching 10.1 g/L [75]. Li, et al. controlled the metabolic pathway of R. eutropha H16 to synthesize fatty acids using H2, CO2, and O2 as substrates, in a gas autotrophic fermentation system. The engineered bacteria B2 generated free fatty acids and reached 60.64 mg/g in less than 48 hours [116]. Hu, et al. modified Clostridium acetogenes to produce biodiesel from synthetic gas in an integrated continuous reactor system with an output of 18 g/L of C16-C18 triacylglycerides [117].
Bioplastics
The production and consumption of non-biodegradable plastics have been steadily increasing over the last few decades, causing a significant environmental load on the environment. The replacement of petroleum-based plastics, on the other hand, offers promising prospects owing to the easy degradability and sustainability of bioplastics. Polyhydroxybutyrate (PHB), is a biodegradable polymer synthesized through microbial fermentation [118]. Proteobacteria, cyanobacteria, purple non-sulfur bacteria, and other microbes produce PHB by a process that is catalyzed by the three enzymes PhaA, PhaB, and PhaC. Mozumder, et al. utilized C. necator in syngas (10% CO2, 75% H2, and 15% O2) to create PHB, with a yield of 42.9 g/L [119]. Karmann et al., designed R. rubrum for synthesizing PHB by utilizing syngas consisting of CO and CO2 as carbon sources and energy sources [120]. Weiss, et al. developed a synthetic symbiotic system where the S. elongatus PCC 7942 CscB secretes sucrose to support Halomonas boliviensis to produce PHB, resulting in a yield of 28.3mg PHB/L/d [121]. Chen et al., constructed a microbial electrosynthesis system (MES) for the CO2-driven synthesis of PHB by R. eutropha [122]. Costa, et al. used a two-stage fermentation with Clostridium autoethanogenum as the host to create PHA [123]. Even though the production of PHB by microorganisms on a large scale has been accomplished using CO2, further research discovered that utilizing sugar as the substrate rather than CO2 as the substrate produced more PHB. Therefore, the ability of microbes to capture and use CO2 still needs significant improvement.
Terpenoids
Terpenoids, referred to as isoprenoids or terpenes, are a broad and diverse class of chemical molecules with several industrial uses, such as in the food, cosmetics, and pharmaceutical industries. Microorganisms produce isoprenoids through the mevalonate pathway (MEV) and methylerythritol 4-phosphate (MEP) pathway [124]. By expressing menthollimonene synthase and abies α-red myrrh synthase, Fiona, et al. effectively engineered Synechococcus sp. PCC7002 to generate limonene and α-red myrrh in polychlorella, with yields of 4 mg/L and 0.6 mg/L, respectively. This achievement offers a promising platform for the production of terpene compounds using algae [89]. The engineered Anabaena sp. PCC7120 was constructed with coexpressed limonene synthase gene (lims) as well as a DXP operon. Under higher light intensity, the limonene yield and productivity increased 6.8 and 8.8 times more than the control strain, respectively [125]. Gao, et al. modified S. elongates to use the methylerythritol phosphate route to produce isoprene, resulting in the designed strain isoprene production reaching 1.26 g/l from CO2 [126].
Hydrocarbons
Hydrocarbons, which are made of carbon and hydrogen, are crucial parts of petroleum. Alkanes and alkenes can be produced by cyanobacteria using a natural carbon fixation route. By optimizing the expression level of the Pseudomonas syringae ethylene forming enzyme (efe code), as well as adjusting light intensity and nutritional conditions, Justin, et al. increased ethylene synthesis in Synechocystis sp. PCC6803 and achieved a maximum ethylene yield of 171 mg/L/d [127].In order to produce ethylene utilizing 2% CO2 as a carbon source, Tomas, et al. overexpressed the Sy-efe gene in Synechocystis sp. PCC 6803 [128].
Biomass
Heterotrophs exclusively consume organic chemicals, while autotrophs possess the ability to utilize CO2 for the synthesis of valuable compounds. The primary objective in synthetic biology is to engineer heterotrophic organisms capable of harnessing CO2 as a carbon source for biomass production. In an electrochemical-biological system designed by Zheng, et al. [103], yeast with a deactivated glucose metabolic pathway produced long carbon chain molecules like glucose and fatty acids from acetic acid and acetate created by electrocatalyzed CO2. This process yielded 2.2 g/L of glucose. Wang et al., genetically modified C.necator to fix CO2 and produce glucose with a yield of 253.3 mg/L, providing a viable approach for microorganisms to generate glucose from CO2 [129].
The bioconversion of CO2 has many advantages, such as low energy consumption, a wide range of products, higher conversion rates in large-scale production, and no competition for food and land resources (Table 2). Around 11.5 million tons of CO2 are annually converted through biotechnology into a variety of goods worldwide, but this number is considerably insufficient in comparison with the annual CO2 emissions (24 billion tons). The application of microorganisms, particularly heterotrophs, to convert CO2 and use it to synthesize bioproducts still needs further research.
| Table 2: Some value-added bioproducts derived from CO2 | |||||
| Pathway | Organism | Carbon Source | Key genes | Product | Ref |
| CBB | E. coli | L-arabinose, xylose | cbbM, PrkA | Ethanol, acetate | [61] |
| CBB | Saccharomyces cerevisiae | maltose, xylose | XR, XDH, XKS, sPRK, cbbM | Ethanol | [64] |
| CBB | S. cerevisiae | glucose, galactose | GroEL, GroES, cbbM, PRK | Ethanol | [63] |
| CBB | S. cerevisiae | glucose, formate | cbbM, prk, fdh | Free fatty acids | [75] |
| CBB | S. elongatus PCC7942 | CO2 | thl, atoAD, adc, adh | Isopropanol | [85] |
| CBB glycolytic |
S. elongatus PCC7942 | CO2, glucose | galp, zwf, gnd, rbcLXS | Butanediol | [86] |
| CBB | S. elongatus PCC7942 | CO2 | Kivd, YqhD | 2-Methyl butanol | [87] |
| CBB | S. elongatus PCC7942 | CO2 | MsLS | Limonene | [88] |
| CBB | S. elongatus PCC7942 | CO2 | pdc, , atfA, xpkA, pta | Fatty acid ethyl esters | [89] |
| CBB | S. elongatus PCC7942 | CO2 | pal | Olefins, Cinnamaldehyde, Curcumin | [90] |
| CBB glycolytic |
E. coli | CO2, glucose | pck, mdh | Malate | [104] |
| CBB | M. extorquens AM1 | CO2 | Prk, cbbM | Cell growth | [34] |
| CBB | C. necator | CO2 | HAD1, cbbY2 | Glucose | [129] |
| CBB | Synechocystis sp. PCC6803 | CO2 | maqu_2220 | Fatty alcohol | [107] |
| CBB | R.eutropha H16 | CO2 | AlsS, ilvCD, kivd & yqhD | Isobutanol, methyl-butanol | [108] |
| CBB | S. cerevisiae | glucose | Rubisco, GroEL/GroES, PRK, non‑ox PPP | Ethanol | [109] |
| CBB | Synechocystis sp. PCC6803 | CO2 | mcr, accB, accC, accA, accD, birA, pntA, pntB | 3-HP | [111] |
| CBB | S. elongatus PCC7942 | CO2 | mgsA | Lactate | [112] |
| CBB rTCA |
R. eutropha H16 | CO2/O2/H2 | acc, Ltes, Fas, acpS | Fatty acids | [116] |
| CBB | C. necator | CO2/O2/H2 | Lipid | [119] | |
| CBB | S. elongatus PCC7942, H. boliviensis |
CO2 | CscB | Ethanol or hydrocarbon fuels | [121] |
| CBB TCA |
C.necator H16 | Glucose, Glycerol, CO2 | ScIPS, EcIMP | myo-inositol | [94] |
| CBB | C. necator H16 | CO2 | bld, adhE, dra, s-adh, PDC | (R)-1,3-butanediol | [95] |
| CBB TCA EM-CoA |
R. rubrum | CO2, acetate | PHB | [120] | |
| TCA | S. cescerevisiae | xylose | PYC1, PYC2, MDH3, SpMAE1 | Malate | [38] |
| TCA | S. cescerevisiae | glucose | RoPYC, SFC1 | fumaric acid | [68] |
| TCA | S. cescerevisiae | Glycerol | PYC2, MDH3-R, fumR, FRDg-R | Succinic acid | [67] |
| TCA | Synechocystis 6803 | CO2 | efe | Ethylene | [127] |
| TCA | E. coli | CO2, glucose | sbtA, bicA, ppc, pck | Succinate | [110] |
| rGly | Methanol, CO2 | Glycine, Serine, Pyruvate | [49] | ||
| rGly | E. coli | CO2, FA | reconstructed THF cycle, gcvTHP, lpd | Glycine, serine | [50] |
| rGly | C. necator | Formate | ftl, fch, mtdA,gcvT, gcvH, gcvP | Biomass | [97] |
| W-L | C. carboxidivorans | CO/CO2/H2 | aor, adhE2, fnr | Ethanol, butanol | [99] |
| W-L | C. ljungdahlii | CO/CO2 | Att/Int system | Butyric acid | [100] |
| W-L | C. ljungdahlii | H2/CO/CO2 | thl, ctfAB, adc | acetone | [101] |
| W-L | M. thermoacetica, Yarrowia lipolytica |
H2/CO2 | Lipid | [117] | |
| W-L | Clostridium autoethanogenum | CO/CO2/H2/N2 | PHA, bioethanol | [123] | |
| W-L | Clostridium acetobutylicum | CO2 | Acetone, butanol | [133] | |
| Electrosynthesis | Clostridium autoethanogenum |
CO2 | Butyrate | [113] | |
| Biosystem-electro | S. cerevisiae | CO2 | yihx, agpP | Long-chain compounds | [103] |
Through synthetic biology and metabolic engineering, the researchers have synthesized carbon sequestration pathways with lower energy consumption and fewer reaction steps, screened enzymes for carbon sequestration with higher carboxylation efficiency, and improved the CO2 assimilation ability of natural CO2-fixing microorganisms, these advancements have further facilitated the achievement of carbon sequestration and emission reduction targets. However, microbial CO2 fixation research is still in the early stages of development, there remain several drawbacks, such as low carbon sequestration efficiency and energy capture efficiency, as well as huge disparities between goal product production and industrial requirements. Therefore, more research is needed to explore in terms of CO2 fixation efficiency, and utilization of energy [130-132].
Thoroughly investigated and optimized natural carbon sequestration pathway and directed evolution of carbon fixation enzymes by rational and semi-rational design enable increase in the efficiency of key enzymes and metabolic modules of the carbon sequestration pathway. In order to efficiently use energy, realize optical drive, and electric drive biocatalysis, it is necessary to optimize the energy utilization systems of electric autotrophs and chemoautotrophs and develop new materials for capturing light energy, generating electricity. Furthermore, by enhancing the electron transfer mechanism and coupling electro-biological CO2 fixation, this will have the potential to use microorganisms on a broad scale to produce added value products from CO2, while simultaneously decreasing the cost of microbial conversion technology.
Financial support from the Nutrition & Health Research Institute, China National Cereals, Oils, and Foodstuffs Corporation (COFCO), and the University of Shihezi is gratefully acknowledged.
- Gupta R, Mishra A, Thirupathaiah Y, Chandel Ak. Biochemical conversion of CO2 in fuels and chemicals: status, innovation, and industrial aspects. Biomass Convers Biorefin. 2022; 1-24.
- Sanna A, Hall MR, Maroto-valer M. Post-processing pathways in carbon capture and storage by mineral carbonation (CCSM) towards the introduction of carbon neutral materials. Energy & Environmental Science. 2012;5:7781.
- Chen L, Msigwa G, Yang M, Osman AI, Fawzy S, Rooney DW, Yap PS. Strategies to achieve a carbon neutral society: a review. Environ Chem Lett. 2022;20(4):2277-2310. doi: 10.1007/s10311-022-01435-8. Epub 2022 Apr 8. PMID: 35431715; PMCID: PMC8992416.
- Kajla S, Kumari R, Nagi GK. Microbial CO2 fixation and biotechnology in reducing industrial CO2 emissions. Arch Microbiol. 2022 Jan 21;204(2):149. doi: 10.1007/s00203-021-02677-w. PMID: 35061105.
- Jaisan C, An DS, Lee DS. Application of Physical Gas Absorbers in Manipulating the CO2 Pressure of Kimchi Package. J Food Sci. 2018 Dec;83(12):3002-3008. doi: 10.1111/1750-3841.14383. Epub 2018 Nov 12. PMID: 30419149.
- Oko E, Ramshaw C, Wang M. Study of intercooling for rotating packed bed absorbers in intensified solvent-based CO2 capture process. Applied Energy. 2018; 223:302-316.
- Mohseni MM, Jouyandeh M, Sajadi SM, Hejna A, Habibzadeh S. Metal-organic frameworks (MOF) based heat transfer: A comprehensive review. Chem Eng J. 2022.
- Ding M , Flaig RW , Jiang HL , Yaghi OM . Carbon capture and conversion using metal-organic frameworks and MOF-based materials. Chem Soc Rev. 2019 May 20;48(10):2783-2828. doi: 10.1039/c8cs00829a. PMID: 31032507.
- Feng J, Zeng S, Feng J, Dong H, Zhang X. CO2 Electroreduction in Ionic Liquids: A Review. Chinese Journal of Chemistry. 2018; 36(10).
- Jones MB, Albanito F. Can biomass supply meet the demands of bioenergy with carbon capture and storage (BECCS)? Glob Chang Biol. 2020 Oct;26(10):5358-5364. doi: 10.1111/gcb.15296. Epub 2020 Aug 20. PMID: 32726492.
- Luis P, Gerven TV, Der Bruggen BV. Recent developments in membrane-based technologies for CO2 capture. Prog Energy Combust Sci. 2012.
- Mulakhudair AR, Al-Mashhadani M, Hanotu J, Zimmerman W. Inactivation combined with cell lysis of Pseudomonas putida using a low pressure carbon dioxide microbubble technology. J Chem Technol Biotechnol. 2017 Aug;92(8):1961-1969. doi: 10.1002/jctb.5299. Epub 2017 May 12. PMID: 28781404; PMCID: PMC5518213.
- Amani H , Habibey R , Hajmiresmail SJ , Latifi S , Pazoki-Toroudi H , Akhavan O . Antioxidant nanomaterials in advanced diagnoses and treatments of ischemia reperfusion injuries. J Mater Chem B. 2017 Dec 28;5(48):9452-9476. doi: 10.1039/c7tb01689a. Epub 2017 Nov 24. PMID: 32264560.
- Wei J, Ge Q, Yao R, Wen Z, Fang C, Guo L, Xu H, Sun J. Directly converting CO2 into a gasoline fuel. Nat Commun. 2017 May 2;8:15174. doi: 10.1038/ncomms15174. Erratum in: Nat Commun. 2017 Oct 12;8:16170. PMID: 28462925; PMCID: PMC5418575.
- Janes T, Yang Y, Song D. Chemical reduction of CO2 facilitated by C-nucleophiles. Chem Commun (Camb). 2017 Oct 17;53(83):11390-11398. doi: 10.1039/c7cc05978g. PMID: 28972211.
- Zha B, Li C, Li J. Efficient electrochemical reduction of CO2 into formate and acetate in polyoxometalate catholyte with indium catalyst. Journal of Catalysis. 2020; 382:69-76.
- Jia S, Ma X, Sun X, Han B. Electrochemical Transformation of CO2 to Value-Added Chemicals and Fuels. CCS Chemistry. 2022; 4(10):17.
- Galadima A, Muraza O. Catalytic thermal conversion of CO2 into fuels: Perspective and challenges. Renewable Sustainable Energy Rev. 2019; 115:109333.
- Feng K, Wang S, Zhang D, Wang L, Yu Y, Feng K, Li Z, Zhu Z, Li C, Cai M, Wu Z, Kong N, Yan B, Zhong J, Zhang X, Ozin GA, He L. Cobalt Plasmonic Superstructures Enable Almost 100% Broadband Photon Efficient CO2 Photocatalysis. Adv Mater. 2020 Jun;32(24):e2000014. doi: 10.1002/adma.202000014. Epub 2020 May 10. PMID: 32390222.
- Long N, Lee J, KOO KK, Luis P, Moonyong L. Recent Progress and Novel Applications in Enzymatic Conversion of Carbon Dioxide. Energies. 2017; 10:473.
- Sullivan I, Goryachev A, Digdaya IA, Atwater HA, Xiang C. Coupling electrochemical CO2 conversion with CO2 capture. Nature Catalysis. 2022; (1):5.
- Choi KR, Ahn YJ, Lee SY. Bacterial conversion of CO2 to organic compounds. J CO2 Util. 2022; 58:C101929.
- Sheng H, Liu C. Spatial decoupling boosts CO2 electro-biofixation. Nat Catal. 2022; 357–358.
- Zahed MA, Movahed E, Khodayari A, Zanganeh S, Badamaki M. Biotechnology for carbon capture and fixation: Critical review and future directions. J Environ Manage. 2021 Sep 1;293:112830. doi: 10.1016/j.jenvman.2021.112830. Epub 2021 May 26. PMID: 34051533.
- Lee J, Park HJ, Moon M, Lee JS, Min K. Recent progress and challenges in microbial polyhydroxybutyrate (PHB) production from CO2 as a sustainable feedstock: A state-of-the-art review. Bioresour Technol. 2021 Nov;339:125616. doi: 10.1016/j.biortech.2021.125616. Epub 2021 Jul 21. PMID: 34304096..
- Schwander T, Schada von Borzyskowski L, Burgener S, Cortina NS, Erb TJ. A synthetic pathway for the fixation of carbon dioxide in vitro. Science. 2016 Nov 18;354(6314):900-904. doi: 10.1126/science.aah5237. PMID: 27856910; PMCID: PMC5892708.
- Bar-Even A, Noor E, Lewis NE, Milo R. Design and analysis of synthetic carbon fixation pathways. Proc Natl Acad Sci U S A. 2010 May 11;107(19):8889-94. doi: 10.1073/pnas.0907176107. Epub 2010 Apr 21. PMID: 20410460; PMCID: PMC2889323.
- Wu C, Lo J, Urban C, Gao X, Yang B. Acetyl-CoA synthesis through a bicyclic carbon-fixing pathway in gas-fermenting. Nature Synthesis. 2022; 1:615-625.
- Xiao L, Liu G, Gong F, Zhu H, Zhang Y. A Minimized Synthetic Carbon Fixation Cycle. ACS Catalysis. 2021; 12:799-808.
- Calvin M, Benson AA. The Path of Carbon in Photosynthesis. Science. 1948 May 7;107(2784):476-80. doi: 10.1126/science.107.2784.476. PMID: 17760010.
- Antonovsky N, Gleizer S, Milo R. Engineering carbon fixation in E. coli: from heterologous RuBisCO expression to the Calvin-Benson-Bassham cycle. Curr Opin Biotechnol. 2017 Oct;47:83-91. doi: 10.1016/j.copbio.2017.06.006. Epub 2017 Jul 15. PMID: 28715702.
- Gleizer S, Ben-Nissan R, Bar-On YM, Antonovsky N, Noor E, Zohar Y, Jona G, Krieger E, Shamshoum M, Bar-Even A, Milo R. Conversion of Escherichia coli to Generate All Biomass Carbon from CO2. Cell. 2019 Nov 27;179(6):1255-1263.e12. doi: 10.1016/j.cell.2019.11.009. PMID: 31778652; PMCID: PMC6904909.
- Gassler T, Sauer M, Gasser B, Egermeier M, Troyer C. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat Biotechnol. 2019; 38:210-216.
- Schada von Borzyskowski L, Carrillo M, Leupold S, Glatter T, Kiefer P, Weishaupt R, Heinemann M, Erb TJ. An engineered Calvin-Benson-Bassham cycle for carbon dioxide fixation in Methylobacterium extorquens AM1. Metab Eng. 2018 May;47:423-433. doi: 10.1016/j.ymben.2018.04.003. Epub 2018 Apr 4. PMID: 29625224.
- Wicker RJ, Kumar G, Khan E, Bhatnagar A. Emergent green technologies for cost-effective valorization of microalgal biomass to renewable fuel products under a biorefinery scheme. Chemical Engineering Journal. 2021; 415:128932.
- Evans MC, Buchanan BB, Arnon DI. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc Natl Acad Sci U S A. 1966 Apr;55(4):928-34. doi: 10.1073/pnas.55.4.928. PMID: 5219700; PMCID: PMC224252.
- Malubhoy Z, Bahia FM, de Valk SC, de Hulster E, Rendulić T, Ortiz JPR, Xiberras J, Klein M, Mans R, Nevoigt E. Carbon dioxide fixation via production of succinic acid from glycerol in engineered Saccharomyces cerevisiae. Microb Cell Fact. 2022 May 28;21(1):102. doi: 10.1186/s12934-022-01817-1. PMID: 35643577; PMCID: PMC9148483.
- Kang NK, Lee JW, Ort DR, Jin YS. L-malic acid production from xylose by engineered Saccharomyces cerevisiae. Biotechnol J. 2022 Mar;17(3):e2000431. doi: 10.1002/biot.202000431. Epub 2021 Aug 25. PMID: 34390209.
- Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta. 2008 Dec;1784(12):1873-98. doi: 10.1016/j.bbapap.2008.08.012. Epub 2008 Aug 27. PMID: 18801467; PMCID: PMC2646786.
- Fast AG, Papoutsakis ET. Functional Expression of the Clostridium ljungdahlii Acetyl-Coenzyme A Synthase in Clostridium acetobutylicum as Demonstrated by a Novel In Vivo CO Exchange Activity En Route to Heterologous Installation of a Functional Wood-Ljungdahl Pathway. Appl Environ Microbiol. 2018 Mar 19;84(7):e02307-17. doi: 10.1128/AEM.02307-17. PMID: 29374033; PMCID: PMC5861816.
- Hu G, Li Z, Ma D, Ye C, Zhang L. Light-driven CO2 sequestration in Escherichia coli to achieve theoretical yield of chemicals. Nature Catalysis. 2021; 4:395-406.
- Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007 Dec 14;318(5857):1782-6. doi: 10.1126/science.1149976. PMID: 18079405.
- Keller MW, Schut GJ, Lipscomb GL, Menon AL, Iwuchukwu IJ, Leuko TT, Thorgersen MP, Nixon WJ, Hawkins AS, Kelly RM, Adams MW. Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):5840-5. doi: 10.1073/pnas.1222607110. Epub 2013 Mar 25. PMID: 23530213; PMCID: PMC3625313.
- Ramos-Vera WH, Berg IA, Fuchs G. Autotrophic carbon dioxide assimilation in Thermoproteales revisited. J Bacteriol. 2009 Jul;191(13):4286-97. doi: 10.1128/JB.00145-09. Epub 2009 May 1. PMID: 19411323; PMCID: PMC2698501.
- Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol. 2011;65:631-58. doi: 10.1146/annurev-micro-090110-102801. PMID: 21740227.
- Liu Z, Wang K, Chen Y, Tan T, Nielsen J. Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nature Catalysis. 2020; 3:274-288.
- Mattozzi Md, Ziesack M, Voges MJ, Silver PA, Way JC. Expression of the sub-pathways of the Chloroflexus aurantiacus 3-hydroxypropionate carbon fixation bicycle in E. coli: Toward horizontal transfer of autotrophic growth. Metab Eng. 2013 Mar;16:130-9. doi: 10.1016/j.ymben.2013.01.005. Epub 2013 Jan 29. PMID: 23376595.
- Claassens NJ. Reductive Glycine Pathway: A Versatile Route for One-Carbon Biotech. Trends Biotechnol. 2021 Apr;39(4):327-329. doi: 10.1016/j.tibtech.2021.02.005. Epub 2021 Feb 23. PMID: 33632541.
- Liu J, Zhang H, Xu Y, Meng H, Zeng AP. Turn air-captured CO2 with methanol into amino acid and pyruvate in an ATP/NAD(P)H-free chemoenzymatic system. Nat Commun. 2023 May 15;14(1):2772. doi: 10.1038/s41467-023-38490-w. PMID: 37188719; PMCID: PMC10185560.
- Bang J, Lee SY. Assimilation of formic acid and CO2 by engineered Escherichia coli equipped with reconstructed one-carbon assimilation pathways. Proc Natl Acad Sci U S A. 2018 Oct 2;115(40):E9271-E9279. doi: 10.1073/pnas.1810386115. Epub 2018 Sep 17. PMID: 30224468; PMCID: PMC6176599.
- Döring V, Darii E, Yishai O, Bar-Even A, Bouzon M. Implementation of a Reductive Route of One-Carbon Assimilation in Escherichia coli through Directed Evolution. ACS Synth Biol. 2018 Sep 21;7(9):2029-2036. doi: 10.1021/acssynbio.8b00167. Epub 2018 Aug 23. PMID: 30106273.
- Gonzalez de la Cruz J, Machens F, Messerschmidt K, Bar-Even A. Core Catalysis of the Reductive Glycine Pathway Demonstrated in Yeast. ACS Synth Biol. 2019 May 17;8(5):911-917. doi: 10.1021/acssynbio.8b00464. Epub 2019 Apr 24. PMID: 31002757; PMCID: PMC6528164.
- Kim S, Lindner SN, Aslan S, Yishai O, Wenk S, Schann K, Bar-Even A. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat Chem Biol. 2020 May;16(5):538-545. doi: 10.1038/s41589-020-0473-5. Epub 2020 Feb 10. PMID: 32042198.
- Köpke M. Redesigning CO2 fixation. Nature Synthesis, 2022; 1:584-585.
- Andersson I. Catalysis and regulation in Rubisco. J Exp Bot. 2008;59(7):1555-68. doi: 10.1093/jxb/ern091. Epub 2008 Apr 15. PMID: 18417482.
- Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot. 2008;59(7):1515-24. doi: 10.1093/jxb/erm361. Epub 2008 Feb 16. PMID: 18281717.
- Pang JJ, Shin JS, Li SY. The Catalytic Role of RuBisCO for in situ CO2 Recycling in Escherichia coli. Front Bioeng Biotechnol. 2020 Nov 30;8:543807. doi: 10.3389/fbioe.2020.543807. PMID: 33330409; PMCID: PMC7734965.
- Fujihashi M, Nishitani Y, Kiriyama T, Aono R, Sato T, Takai T, Tagashira K, Fukuda W, Atomi H, Imanaka T, Miki K. Mutation design of a thermophilic Rubisco based on three-dimensional structure enhances its activity at ambient temperature. Proteins. 2016 Oct;84(10):1339-46. doi: 10.1002/prot.25080. Epub 2016 Jun 24. PMID: 27273261.
- Aigner H, Wilson RH, Bracher A, Calisse L, Bhat JY, Hartl FU, Hayer-Hartl M. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science. 2017 Dec 8;358(6368):1272-1278. doi: 10.1126/science.aap9221. PMID: 29217567.
- Zhuang ZY, Li SY. Rubisco-based engineered Escherichia coli for in situ carbon dioxide recycling. Bioresour Technol. 2013 Dec;150:79-88. doi: 10.1016/j.biortech.2013.09.116. Epub 2013 Oct 3. PMID: 24152790.
- Xia PF, Zhang GC, Walker B, Seo SO, Kwak S, Liu JJ, Kim H, Ort DR, Wang SG, Jin YS. Recycling Carbon Dioxide during Xylose Fermentation by Engineered Saccharomyces cerevisiae. ACS Synth Biol. 2017 Feb 17;6(2):276-283. doi: 10.1021/acssynbio.6b00167. Epub 2016 Oct 31. PMID: 27744692.
- Guadalupe-Medina V, Wisselink HW, Luttik MA, de Hulster E, Daran JM, Pronk JT, van Maris AJ. Carbon dioxide fixation by Calvin-Cycle enzymes improves ethanol yield in yeast. Biotechnol Biofuels. 2013 Aug 29;6(1):125. doi: 10.1186/1754-6834-6-125. PMID: 23987569; PMCID: PMC3766054.
- Li YJ, Wang MM, Chen YW, Wang M, Fan LH, Tan TW. Engineered yeast with a CO2-fixation pathway to improve the bio-ethanol production from xylose-mixed sugars. Sci Rep. 2017 Mar 6;7:43875. doi: 10.1038/srep43875. PMID: 28262754; PMCID: PMC5338314.
- Matsumura H, Shiomi K, Yamamoto A, Taketani Y, Kobayashi N, Yoshizawa T, Tanaka SI, Yoshikawa H, Endo M, Fukayama H. Hybrid Rubisco with Complete Replacement of Rice Rubisco Small Subunits by Sorghum Counterparts Confers C4 Plant-like High Catalytic Activity. Mol Plant. 2020 Nov 2;13(11):1570-1581. doi: 10.1016/j.molp.2020.08.012. Epub 2020 Aug 31. PMID: 32882392.
- Rin Kim S, Kim SJ, Kim SK, Seo SO, Park S, Shin J, Kim JS, Park BR, Jin YS, Chang PS, Park YC. Yeast metabolic engineering for carbon dioxide fixation and its application. Bioresour Technol. 2022 Feb;346:126349. doi: 10.1016/j.biortech.2021.126349. Epub 2021 Nov 17. PMID: 34800639.
- Zelle RM, de Hulster E, van Winden WA, de Waard P, Dijkema C, Winkler AA, Geertman JM, van Dijken JP, Pronk JT, van Maris AJ. Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl Environ Microbiol. 2008 May;74(9):2766-77. doi: 10.1128/AEM.02591-07. Epub 2008 Mar 14. PMID: 18344340; PMCID: PMC2394876.
- Xiberras J, Klein M, de Hulster E, Mans R, Nevoigt E. Engineering Saccharomyces cerevisiae for Succinic Acid Production From Glycerol and Carbon Dioxide. Front Bioeng Biotechnol. 2020 Jun 26;8:566. doi: 10.3389/fbioe.2020.00566. PMID: 32671027; PMCID: PMC7332542.
- Xu G, Zou W, Chen X, Xu N, Liu L, Chen J. Fumaric acid production in Saccharomyces cerevisiae by in silico aided metabolic engineering. PLoS One. 2012;7(12):e52086. doi: 10.1371/journal.pone.0052086. Epub 2012 Dec 26. PMID: 23300594; PMCID: PMC3530589.
- Talekar S, Jo BH, Dordick JS, Kim J. Carbonic anhydrase for CO2 capture, conversion and utilization. Curr Opin Biotechnol. 2022 Apr;74:230-240. doi: 10.1016/j.copbio.2021.12.003. Epub 2022 Jan 3. PMID: 34992045.
- Gong F, Liu G, Zhai X, Zhou J, Cai Z, Li Y. Quantitative analysis of an engineered CO2-fixing Escherichia coli reveals great potential of heterotrophic CO2 fixation. Biotechnol Biofuels. 2015 Jun 18;8:86. doi: 10.1186/s13068-015-0268-1. PMID: 26097503; PMCID: PMC4475311.
- Sri Wahyu Effendi S, Lin JY, Ng IS. Simultaneous carbon dioxide sequestration and utilization for cadaverine production using dual promoters in engineered Escherichia coli strains. Bioresour Technol. 2022 Nov;363:127980. doi: 10.1016/j.biortech.2022.127980. Epub 2022 Sep 19. PMID: 36137445.
- Moon M, Park GW, Lee JP, Lee JS, Min K. Recombinant expression and characterization of formate dehydrogenase from Clostridium ljungdahlii (ClFDH) as CO2 reductase for converting CO2 to formate. J. CO2 Util. 2022; 57:101876.
- Aresta M, Dibenedetto A. Utilisation of CO2 as a chemical feedstock: opportunities and challenges. Dalton Trans. 2007 Jul 28;(28):2975-92. doi: 10.1039/b700658f. Epub 2007 Jun 26. PMID: 17622414.
- Du C, Li Y, He Y, Su L, Wang H. Fixing carbon dioxide in situ during ethanol production by formate dehydrogenase. Green Chemistry. 2022; 24:6989-6999.
- Wang K, Da Y, Bi H, Liu Y, Chen B. A one-carbon chemicals conversion strategy to produce precursor of biofuels with Saccharomyces cerevisiae. Renewable Energy. 2023; 208:331-340.
- Roger M, Reed TCP, Sargent F. Harnessing Escherichia coli for Bio-Based Production of Formate under Pressurized H2 and CO2 Gases. Appl Environ Microbiol. 2021 Oct 14;87(21):e0029921. doi: 10.1128/AEM.00299-21. Epub 2021 Sep 8. PMID: 34647819; PMCID: PMC8516059.
- Roger M, Brown F, Gabrielli W, Sargent F. Efficient Hydrogen-Dependent Carbon Dioxide Reduction by Escherichia coli. Curr Biol. 2018 Jan 8;28(1):140-145.e2. doi: 10.1016/j.cub.2017.11.050. Epub 2017 Dec 28. PMID: 29290558; PMCID: PMC5772173.
- Choi ES, Min K, Kim GJ, Kwon I, Kim YH. Expression and characterization of Pantoea CO dehydrogenase to utilize CO-containing industrial waste gas for expanding the versatility of CO dehydrogenase. Sci Rep. 2017 Mar 14;7:44323. doi: 10.1038/srep44323. PMID: 28290544; PMCID: PMC5349547.
- Jin X, Gong S, Chen Z, Xia J, Xiang W. Potential microalgal strains for converting flue gas CO2 into biomass. J. Appl. Phycol. 2020; 33:47-55.
- Singh HM, Kothari R, Gupta R, Tyagi VV. Bio-fixation of flue gas from thermal power plants with algal biomass: Overview and research perspectives. J Environ Manage. 2019 Sep 1;245:519-539. doi: 10.1016/j.jenvman.2019.01.043. Epub 2019 Feb 23. PMID: 30803750.
- Lee TM, Lin JY, Tsai TH, Yang RY, Ng IS. Clustered regularly interspaced short palindromic repeats (CRISPR) technology and genetic engineering strategies for microalgae towards carbon neutrality: A critical review. Bioresour Technol. 2023 Jan;368:128350. doi: 10.1016/j.biortech.2022.128350. Epub 2022 Nov 19. PMID: 36414139.
- Wei L, Wang Q, Xin Y, Lu Y, Xu J. Enhancing photosynthetic biomass productivity of industrial oleaginous microalgae by overexpression of RuBisCO activase. Algal Research. 2017; 27:366-375.
- Wang W, Yu LJ, Xu C, Tomizaki T, Zhao S, Umena Y, Chen X, Qin X, Xin Y, Suga M, Han G, Kuang T, Shen JR. Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science. 2019 Feb 8;363(6427):eaav0365. doi: 10.1126/science.aav0365. PMID: 30733387.
- Shin WS, Lee B, Jeong BR, Chang YK, Kwon JH. Truncated light-harvesting chlorophyll antenna size in Chlorella vulgaris improves biomass productivity. J. Appl. Phycol. 2016; 28:3193-3202.
- Kusakabe T, Tatsuke T, Tsuruno K, Hirokawa Y, Atsumi S, Liao JC, Hanai T. Engineering a synthetic pathway in cyanobacteria for isopropanol production directly from carbon dioxide and light. Metab Eng. 2013 Nov;20:101-8. doi: 10.1016/j.ymben.2013.09.007. Epub 2013 Sep 25. PMID: 24076145.
- Kanno M, Carroll AL, Atsumi S. Global metabolic rewiring for improved CO2 fixation and chemical production in cyanobacteria. Nat Commun. 2017 Mar 13;8:14724. doi: 10.1038/ncomms14724. PMID: 28287087; PMCID: PMC5355792.
- Shen CR, Liao JC. Photosynthetic production of 2-methyl-1-butanol from CO2 in cyanobacterium Synechococcus elongatus PCC7942 and characterization of the native acetohydroxyacid synthase. Energy & Environmental Science. 2012; 5; 9574.
- Davies FK, Work VH, Beliaev AS, Posewitz MC. Engineering Limonene and Bisabolene Production in Wild Type and a Glycogen-Deficient Mutant of Synechococcus sp. PCC 7002. Front Bioeng Biotechnol. 2014 Jun 19;2:21. doi: 10.3389/fbioe.2014.00021. PMID: 25152894; PMCID: PMC4126464.
- Lee HJ, Choi J, Lee SM, Um Y, Sim SJ, Kim Y, Woo HM. Photosynthetic CO2 Conversion to Fatty Acid Ethyl Esters (FAEEs) Using Engineered Cyanobacteria. J Agric Food Chem. 2017 Feb 15;65(6):1087-1092. doi: 10.1021/acs.jafc.7b00002. Epub 2017 Feb 2. PMID: 28128561.
- Li C, Yin L, Wang J, Zheng H, Ni J. Light-driven biosynthesis of volatile, unstable and photosensitive chemicals from CO2. Nature Synthesis. 2023.
- Park JY, Kim BN, Kim YH, Min J. Whole-genome sequence of purple non-sulfur bacteria, Rhodobacter sphaeroides strain MBTLJ-8 with improved CO2 reduction capacity. J Biotechnol. 2018 Dec 20;288:9-14. doi: 10.1016/j.jbiotec.2018.10.007. Epub 2018 Oct 22. Erratum in: J Biotechnol. 2019 Jan 20;290:67. PMID: 30359676.
- Fixen KR, Zheng Y, Harris DF, Shaw S, Yang ZY, Dean DR, Seefeldt LC, Harwood CS. Light-driven carbon dioxide reduction to methane by nitrogenase in a photosynthetic bacterium. Proc Natl Acad Sci U S A. 2016 Sep 6;113(36):10163-7. doi: 10.1073/pnas.1611043113. Epub 2016 Aug 22. PMID: 27551090; PMCID: PMC5018805.
- Panich J, Fong B, Singer SW. Metabolic Engineering of Cupriavidus necator H16 for Sustainable Biofuels from CO2. Trends Biotechnol. 2021 Apr;39(4):412-424. doi: 10.1016/j.tibtech.2021.01.001. Epub 2021 Jan 29. PMID: 33518389.
- Wang X, Wang K, Wang L, Luo H, Wang Y, Wang Y, Tu T, Qin X, Su X, Bai Y, Yao B, Huang H, Zhang J. Engineering Cupriavidus necator H16 for heterotrophic and autotrophic production of myo-inositol. Bioresour Technol. 2023 Jan;368:128321. doi: 10.1016/j.biortech.2022.128321. Epub 2022 Nov 13. PMID: 36379295.
- Gascoyne JL, Bommareddy RR, Heeb S, Malys N. Engineering Cupriavidus necator H16 for the autotrophic production of (R)-1,3-butanediol. Metab Eng. 2021 Sep;67:262-276. doi: 10.1016/j.ymben.2021.06.010. Epub 2021 Jul 2. PMID: 34224897; PMCID: PMC8449065.
- Liu C, Colón BC, Ziesack M, Silver PA, Nocera DG. Water splitting-biosynthetic system with CO₂ reduction efficiencies exceeding photosynthesis. Science. 2016 Jun 3;352(6290):1210-3. doi: 10.1126/science.aaf5039. PMID: 27257255.
- Claassens NJ, Bordanaba-Florit G, Cotton CAR, De Maria A, Finger-Bou M, Friedeheim L, Giner-Laguarda N, Munar-Palmer M, Newell W, Scarinci G, Verbunt J, de Vries ST, Yilmaz S, Bar-Even A. Replacing the Calvin cycle with the reductive glycine pathway in Cupriavidus necator. Metab Eng. 2020 Nov;62:30-41. doi: 10.1016/j.ymben.2020.08.004. Epub 2020 Aug 15. PMID: 32805426.
- Pan H, Wang J, Wu H, Li Z, Lian J. Synthetic biology toolkit for engineering Cupriviadus necator H16 as a platform for CO2 valorization. Biotechnol Biofuels. 2021 Nov 4;14(1):212. doi: 10.1186/s13068-021-02063-0. PMID: 34736496; PMCID: PMC8570001.
- Cheng C, Li W, Lin M, Yang ST. Metabolic engineering of Clostridium carboxidivorans for enhanced ethanol and butanol production from syngas and glucose. Bioresour Technol. 2019 Jul;284:415-423. doi: 10.1016/j.biortech.2019.03.145. Epub 2019 Mar 30. PMID: 30965197.
- Huang H, Chai C, Yang S, Jiang W, Gu Y. Phage serine integrase-mediated genome engineering for efficient expression of chemical biosynthetic pathway in gas-fermenting Clostridium ljungdahlii. Metab Eng. 2019 Mar;52:293-302. doi: 10.1016/j.ymben.2019.01.005. Epub 2019 Jan 8. PMID: 30633974.
- Jones SW, Fast AG, Carlson ED, Wiedel CA, Au J, Antoniewicz MR, Papoutsakis ET, Tracy BP. CO2 fixation by anaerobic non-photosynthetic mixotrophy for improved carbon conversion. Nat Commun. 2016 Sep 30;7:12800. doi: 10.1038/ncomms12800. PMID: 27687501; PMCID: PMC5056431.
- van Aalst ACA, Mans R, Pronk JT. An engineered non-oxidative glycolytic bypass based on Calvin-cycle enzymes enables anaerobic co-fermentation of glucose and sorbitol by Saccharomyces cerevisiae. Biotechnol Biofuels Bioprod. 2022 Oct 17;15(1):112. doi: 10.1186/s13068-022-02200-3. PMID: 36253796; PMCID: PMC9578259.
- Zheng T, Zhang M, Wu L, Guo S, Liu X. Upcycling CO2 into energy-rich long-chain compounds via electrochemical and metabolic engineering. Nature Catalysis. 2022; 5:388.
- Hu G, Zhou J, Chen X, Qian Y, Gao C, Guo L, Xu P, Chen W, Chen J, Li Y, Liu L. Engineering synergetic CO2-fixing pathways for malate production. Metab Eng. 2018 May;47:496-504. doi: 10.1016/j.ymben.2018.05.007. Epub 2018 May 16. PMID: 29753840.
- Zhang Y, Zhou J, Zhang Y, Liu T, Lu X, Men D, Zhang XE. Auxiliary Module Promotes the Synthesis of Carboxysomes in E. coli to Achieve High-Efficiency CO2 Assimilation. ACS Synth Biol. 2021 Apr 16;10(4):707-715. doi: 10.1021/acssynbio.0c00436. Epub 2021 Mar 16. PMID: 33723997.
- Zhu P, Chen X. Converting heterotrophic Escherichia coli into synthetic C1-trophic modes. Trends in Chemistry. 2022; 4:860-862.
- Yao L, Qi F, Tan X, Lu X. Improved production of fatty alcohols in cyanobacteria by metabolic engineering. Biotechnol Biofuels. 2014 Jun 18;7:94. doi: 10.1186/1754-6834-7-94. PMID: 25024742; PMCID: PMC4096523.
- Li H, Opgenorth PH, Wernick DG, Rogers S, Wu TY, Higashide W, Malati P, Huo YX, Cho KM, Liao JC. Integrated electromicrobial conversion of CO2 to higher alcohols. Science. 2012 Mar 30;335(6076):1596. doi: 10.1126/science.1217643. PMID: 22461604.
- Papapetridis I, Goudriaan M, Vázquez Vitali M, de Keijzer NA, van den Broek M, van Maris AJA, Pronk JT. Optimizing anaerobic growth rate and fermentation kinetics in Saccharomyces cerevisiae strains expressing Calvin-cycle enzymes for improved ethanol yield. Biotechnol Biofuels. 2018 Jan 25;11:17. doi: 10.1186/s13068-017-1001-z. PMID: 29416562; PMCID: PMC5784725.
- Yu JH, Zhu LW, Xia ST, Li HM, Tang YL, Liang XH, Chen T, Tang YJ. Combinatorial optimization of CO2 transport and fixation to improve succinate production by promoter engineering. Biotechnol Bioeng. 2016 Jul;113(7):1531-41. doi: 10.1002/bit.25927. Epub 2016 Feb 3. PMID: 26724788.
- Wang Y, Sun T, Gao X, Shi M, Wu L, Chen L, Zhang W. Biosynthesis of platform chemical 3-hydroxypropionic acid (3-HP) directly from CO2 in cyanobacterium Synechocystis sp. PCC 6803. Metab Eng. 2016 Mar;34:60-70. doi: 10.1016/j.ymben.2015.10.008. Epub 2015 Nov 9. PMID: 26546088.
- Hirokawa Y, Goto R, Umetani Y, Hanai T. Construction of a novel d-lactate producing pathway from dihydroxyacetone phosphate of the Calvin cycle in cyanobacterium, Synechococcus elongatus PCC 7942. J Biosci Bioeng. 2017 Jul;124(1):54-61. doi: 10.1016/j.jbiosc.2017.02.016. Epub 2017 Mar 18. PMID: 28325659.
- Batlle-Vilanova P, Ganigué R, Ramió-Pujol S, Bañeras L, Jiménez G, Hidalgo M, Balaguer MD, Colprim J, Puig S. Microbial electrosynthesis of butyrate from carbon dioxide: Production and extraction. Bioelectrochemistry. 2017 Oct;117:57-64. doi: 10.1016/j.bioelechem.2017.06.004. Epub 2017 Jun 15. PMID: 28633067.
- Salehizadeh H, Yan N, Farnood R. Recent advances in microbial CO2 fixation and conversion to value-added products. Chemical Engineering Journal. 2020; 390:124584.
- Yunus IS, Jones PR. Photosynthesis-dependent biosynthesis of medium chain-length fatty acids and alcohols. Metab Eng. 2018 Sep;49:59-68. doi: 10.1016/j.ymben.2018.07.015. Epub 2018 Jul 25. PMID: 30055323.
- Li Z, Xiong B, Liu L, Li S, Xin X, Li Z, Zhang X, Bi C. Development of an autotrophic fermentation technique for the production of fatty acids using an engineered Ralstonia eutropha cell factory. J Ind Microbiol Biotechnol. 2019 Jun;46(6):783-790. doi: 10.1007/s10295-019-02156-8. Epub 2019 Feb 27. Erratum in: J Ind Microbiol Biotechnol. 2019 Apr 10;: PMID: 30810844.
- Hu P, Chakraborty S, Kumar A, Woolston B, Liu H, Emerson D, Stephanopoulos G. Integrated bioprocess for conversion of gaseous substrates to liquids. Proc Natl Acad Sci U S A. 2016 Apr 5;113(14):3773-8. doi: 10.1073/pnas.1516867113. Epub 2016 Mar 7. PMID: 26951649; PMCID: PMC4833252.
- Sirohi R, Kumar Gaur V, Kumar Pandey A, Jun Sim S, Kumar S. Harnessing fruit waste for poly-3-hydroxybutyrate production: A review. Bioresour Technol. 2021 Apr;326:124734. doi: 10.1016/j.biortech.2021.124734. Epub 2021 Jan 20. PMID: 33497926.
- Islam Mozumder MS, Garcia-Gonzalez L, Wever HD, Volcke Eveline IP. Poly(3-hydroxybutyrate) (PHB) production from CO2: Model development and process optimization. Biochemical Engineering Journal. 2015; 98:107-116.
- Karmann S, Panke S, Zinn M. Fed-Batch Cultivations of Rhodospirillum rubrum Under Multiple Nutrient-Limited Growth Conditions on Syngas as a Novel Option to Produce Poly(3-Hydroxybutyrate) (PHB). Front Bioeng Biotechnol. 2019 Apr 2;7:59. doi: 10.3389/fbioe.2019.00059. PMID: 31001525; PMCID: PMC6454858.
- Weiss TL, Young EJ, Ducat DC. A synthetic, light-driven consortium of cyanobacteria and heterotrophic bacteria enables stable polyhydroxybutyrate production. Metab Eng. 2017 Nov;44:236-245. doi: 10.1016/j.ymben.2017.10.009. Epub 2017 Oct 20. PMID: 29061492.
- Chen X, Cao Y, Li F, Tian Y, Song H. Enzyme-assisted microbial electrosynthesis of poly(3-hydroxybutyrate) via co2 bioreduction by engineered Ralstonia eutropha. ACS Catalysis. 2018; 8:4429-4437.
- Lagoa-Costa B, Abubackar HN, Fernández-Romasanta M, Kennes C, Veiga MC. Integrated bioconversion of syngas into bioethanol and biopolymers. Bioresour Technol. 2017 Sep;239:244-249. doi: 10.1016/j.biortech.2017.05.019. Epub 2017 May 5. PMID: 28521235.
- Frank A, Groll M. The Methylerythritol Phosphate Pathway to Isoprenoids. Chem Rev. 2017 Apr 26;117(8):5675-5703. doi: 10.1021/acs.chemrev.6b00537. Epub 2016 Dec 20. PMID: 27995802.
- Halfmann C, Gu L, Zhou R. Engineering cyanobacteria for the production of a cyclic hydrocarbon fuel from CO2 and H2O. Green Chem. 2014; 16:3175-3185.
- Gao X, Gao F, Liu D, Zhang H, Nie X, Yang C. Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy & Environmental Science. 2016; 9:1400.
- Ungerer J, Tao L, Davis M, Ghirardi M, Maness PC. Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803†. Energy & Environmental Science. 2012; 5:8998.
- Zavřel T, Knoop H, Steuer R, Jones PR, Červený J, Trtílek M. A quantitative evaluation of ethylene production in the recombinant cyanobacterium Synechocystis sp. PCC 6803 harboring the ethylene-forming enzyme by membrane inlet mass spectrometry. Bioresour Technol. 2016 Feb;202:142-51. doi: 10.1016/j.biortech.2015.11.062. Epub 2015 Dec 2. PMID: 26708481.
- Wang X, Luo H, Wang Y, Wang Y, Tu T, Qin X, Su X, Huang H, Bai Y, Yao B, Zhang J. Direct conversion of carbon dioxide to glucose using metabolically engineered Cupriavidus necator. Bioresour Technol. 2022 Oct;362:127806. doi: 10.1016/j.biortech.2022.127806. Epub 2022 Aug 27. PMID: 36031135.
- Tan X, Nielsen J. The integration of bio-catalysis and electrocatalysis to produce fuels and chemicals from carbon dioxide. Chem Soc Rev. 2022 Jun 6;51(11):4763-4785. doi: 10.1039/d2cs00309k. PMID: 35584360.
- Hu L, Guo S, Wang B, Fu R, Fan D, Jiang M, Fei Q, Gonzalez R. Bio-valorization of C1 gaseous substrates into bioalcohols: Potentials and challenges in reducing carbon emissions. Biotechnol Adv. 2022 Oct;59:107954. doi: 10.1016/j.biotechadv.2022.107954. Epub 2022 Apr 10. PMID: 35417775.
- Burlacot A, Dao O, Auroy P, Cuiné S, Li-Beisson Y, Peltier G. Alternative photosynthesis pathways drive the algal CO2-concentrating mechanism. Nature. 2022 May;605(7909):366-371. doi: 10.1038/s41586-022-04662-9. Epub 2022 Apr 27. PMID: 35477755.
- Ampelli C, Perathoner S, Centi G. CO2 utilization: an enabling element to move to a resource- and energy-efficient chemical and fuel production. Philos Trans A Math Phys Eng Sci. 2015 Mar 13;373(2037):20140177. doi: 10.1098/rsta.2014.0177. PMID: 25666059.